
LncRNA SNHG1 correlates with higher T stage and worse overall survival, and promotes cell proliferation while reduces cell apoptosis in breast cancer
Introduction
Breast cancer is the most common cancer and the leading cause of cancer death in females. It is reported that more than one million individuals are diagnosed with breast cancer and over 400,000 females die from this disease every year all around the world (1-3). Despite that the incidence rate of breast cancer in China is relatively low, the increment speed of its incidence in China is much higher than that in western countries (4,5). Recently, great progress on the prognosis has been achieved owning to the improved screening strategies and novel therapeutic approaches for breast cancer, whereas the survival of breast cancer patients remains unfavorable due to the high rate of recurrence and metastasis, especially for triple-negative breast cancer patients (6,7). Therefore, exploring novel biomarkers that are associated with the prognosis of breast cancer patients, and finding novel therapeutic targets are of great value to the management of this disease.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs with more than 200 nucleotides, which participate in a variety of activities and are observed to be dysregulated in diverse cancers (8-13). As one of the members of lncRNA family, the small nucleolar RNA host gene 1 (SNHG1) that locates at 11q12.3 locus is up-regulated in a number of cancers such as cervical cancer, colon cancer and glioma, and it correlates with worse prognosis in cancer patients. Besides, studies find that increased lncRNA SNHG1 expression in these cancer cells enhances cancer cell proliferation, migration and invasion but inhibits cell apoptosis (14-16). We therefore hypothesized that lncRNA SNHG1 is also involved in the development and prognosis of breast cancer. However, no related study has been conducted so far. Hence the aim of this study was to investigate the correlation of lncRNA SNHG1 with prognosis in breast cancer patients, and its effect on breast cancer cell proliferation and apoptosis.
Methods
Patients
A total of 178 breast cancer patients underwent resection at Department of Thyroid and Breast Surgery in The Central Hospital of Wuhan were consecutively enrolled in this study from Jan 2012 to Dec 2013. The inclusion criteria were as follows: (I) patients were diagnosed as primary breast cancer according to clinical, imaging and pathological findings; (II) age above 18 years; (III) about to undergo resection; (IV) able to be followed up regularly. This study was approved by the Ethics Review Board of The Central Hospital of Wuhan (No. 2011-12), all the patients provided signed informed consents.
Sample collection
Tumor tissue and paired adjacent tissue were obtained from breast cancer patients during the operation and immediately stored in the liquid nitrogen for further detection.
LncRNA SNHG1 measurement and clinical assessments
LncRNA SNHG1 level in tumor tissue and adjacent tissue was determined by quantitative polymerase chain reaction (qPCR) assay. Tumor features including molecular subtype, pathological grade, TNM stage, human epidermal growth factor receptor 2 (HER-2), estrogen receptor (ER) and progesterone receptor (PR) status were documented, and their correlation with lncRNA SNHG1 level was further analyzed. In addition, patients were followed up every 1 month in the first half year, and every 3–6 months in the following duration until 48 months (the last follow-up date was 2017/12/31, and a total of 21 patients lost follow up during follow-up period). Finally, all 178 patients were included in the analysis. Overall survival (OS) was calculated, and the comparison of OS between patients with lncRNA SNHG1 high expression and low expression was conducted. As for patients who lost follow up, they were considered as censored data in the analysis of OS.
Cell culture and plasmid transfections
BT474, MCF7, MDA-MB-453, MDA-MB-231, HCC1937 and MCF10A cell lines were purchased from Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China). The former five cell lines were breast cancer cells while the latter one was normal breast epithelial cells. As to the culture condition of these cells, BT474 and HCC1937 cells were cultured in 90% RPMI-1640 sodium (GIBCO, USA) with 10% fetal bovine serum (FBS) (GIBCO, USA); MCF7 cells were cultured in 90% MEM sodium (GIBCO, USA) with 10% FBS (GIBCO, USA); MDA-MB-453 and MDA-MB-231 cells were cultured in 90% L-15 sodium (GIBCO, USA) with 10% FBS (GIBCO, USA); MCF10A cells were cultured in MEGM kit (Sigma, USA). All cells were cultured under 5% CO2 at 37 °C. Afterward, the expression of lncRNA SNHG1 in each cell line was determined by qPCR. Subsequently, we transferred the negative control (NC) overexpression plasmids [NC (+) group], lncRNA SNHG1 overexpression plasmids [LncRNA (+) group], NC short hairpin RNA (shRNA) plasmids [NC (−) group] and lncRNA SNHG1 shRNA plasmids [LncRNA (−) group] into MDA-MB-453 cells and MCF7 cells, and the plasmids were constructed using PET-1 and pGPH1 plasmids (NTCC, China).
Detection of cell proliferation and apoptosis after transfections
After transfection of MDA-MB-453 cells and MCF7 cells, Cell Counting Kit-8 (CCK8) assay was performed at 0, 24, 48 and 72 h to measure cell proliferation using CCK8 commercial kit (R&D, USA), and Annexin V-fluorescein isothiocyanate/propidium iodide staining (AV/PI) assay was performed at 72 h to measure cell apoptosis rate using AV/PI commercial kit (Thermo, USA). In addition, apoptotic and anti-apoptotic markers C-Caspase 3 and p-P38 expressions were also measured at 72 h by Western blot. All the experiments were carried out according to the manufactures’ instructions.
qPCR assay
LncRNA level was determined by qPCR. Total RNA was extracted by TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. Then RNA was quantified by OD 260, and 1 µg of total RNA from each sample was used for cDNA synthesis with QuantiNova Reverse Transcription Kit (Qiagen, German). The cDNA product was subsequently subjected to qPCR with SYBR Green kit (TaKaRa, Japan). PCR amplification was performed as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 s, 61 °C for 30 s. Subsequently, the expressions of candidate lncRNAs were calculated using the 2−ΔΔCt methods with phosphoglyceraldehyde dehydrogenase (GAPDH) as internal reference. The primer of lncRNA SNHG1 was: forward 5'-AGGCTGAAGTTACAGGTC-3', reverse 5'-TTGGCTCCCAGTGTCTTA-3'; the primer of GAPDH was: forward 5'-GAGTCCACTGGCGTCTTCAC-3', reverse 5'-ATCTTGAGGCTGTTGTCATACTTCT-3'.
Western blot assay
Total protein was extracted from MDA-MB-453 cells and MCF7 cells with 1 ml RIPA buffer (Thermo Fisher Scientific, USA) and then its concentration was measured using bicinchoninic acid (BCA) kit (Pierce Biotechnology, USA) and adjudged according to the standard curve, respectively. Subsequently, 20 µg of protein samples with equal concentration were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (Millipore, Bedford, USA). After blocking with 5% skim milk for 2 h, membranes were incubated with the corresponding primary antibody overnight at 4 °C. Then, the membranes were incubated with the appropriate secondary antibody for 1 h at room temperature. The bands were visualized using an enhanced chemiluminescence (ECL) kit (Millipore, Bedford, USA) followed by exposure to X-ray film. Primary antibodies utilized in this study were: cleaved caspase-3 rabbit mAb (CST, USA), dilution 1:1,000; rabbit polyclonal to caspase-3 (CST, USA), dilution 1:500; p38 MAPK antibody (CST, USA), dilution 1:1,000; phospho-p38 MAPK antibody (CST, USA), dilution 1:1,000; rabbit monoclonal to GAPDH (CST, USA), dilution 1:1,000. The secondary antibody used in this study was anti-rabbit IgG, HRP-linked antibody (CST, USA), dilution 1:2,000.
Statistics
The statistical analysis was performed by SPSS 21.0 (IBM, USA) and Graphpad Prism 5.01 software (GraphPad Software Inc, USA). Data were presented as mean ± standard deviation or count (percentage). Comparison between two individual groups was determined by t-test or Chi-square test, while comparison between two paired groups was determined by Wilcoxon signed-rank sum test. Kaplan-Meier (K-M) curves and log-rank test were performed to compare the OS between two groups. P<0.05 was considered significant.
Results
Characteristics of breast cancer patients with high or low lncRNA SNHG1 expression in tumor tissue
Breast cancer patients were divided into lncRNA SNHG1 high expression patients (N=89) and lncRNA SNHG1 low expression patients (N=89) according to the median level of lncRNA SNHG1 in tumor tissues, and T stage was observed to be higher in lncRNA SNHG1 high expression patients compared with lncRNA SNHG1 low expression patients (P=0.001). Whereas no difference was discovered in molecular subtype (P=0.555), pathological grade (P=0.377), N stage (P=0.214), TNM stage (P=0.075), ER status (P=0.539), PR status (P=0.549), HER2 status (P=0.430) or subsequent therapy (all P>0.05) between lncRNA SNHG1 high expression patients and lncRNA SNHG1 low expression patients (Table 1).
Table 1
| Parameters | LncRNA SNHG1 low expression (N=89) | LncRNA SNHG1 high expression (N=89) | P value |
|---|---|---|---|
| Age (years) | 54.39±12.41 | 53.27±12.79 | 0.553 |
| Molecular subtype | |||
| Luminal A | 36 (40.4) | 31 (34.8) | |
| Luminal B | 22 (24.7) | 23 (25.8) | |
| ERBB2+ | 17 (19.1) | 14 (15.7) | |
| Basal-like | 14 (15.7) | 21 (23.6) | |
| Pathological grade (n/%) | |||
| Grade 1 | 18 (20.2) | 21 (23.6) | |
| Grade 2 | 67 (75.3) | 60 (67.4) | |
| Grade 3 | 4 (4.5) | 8 (9.0) | |
| T stage (n/%) | 0.001 | ||
| T1 | 32 (36.0) | 23 (25.8) | |
| T2 | 56 (62.9) | 50 (56.2) | |
| T3 | 1 (1.1) | 16 (18.0) | |
| N stage (n/%) | 0.214 | ||
| N0 | 48 (53.9) | 39 (43.8) | |
| N1 | 26 (29.2) | 28 (31.5) | |
| N2 | 15 (16.9) | 19 (21.3) | |
| N3 | 0 (0.0) | 3 (3.4) | |
| TNM stage (n/%) | 0.075 | ||
| Stage I | 12 (13.5) | 10 (11.2) | |
| Stage II | 62 (69.7) | 51 (57.3) | |
| Stage III | 15 (16.9) | 28 (31.5) | |
| ER status | 0.539 | ||
| Positive | 56 (62.9) | 52 (58.4) | |
| Negative | 33 (37.1) | 37 (41.6) | |
| PR status | 0.549 | ||
| Positive | 47 (52.8) | 43 (48.3) | |
| Negative | 42 (47.2) | 46 (51.7) | |
| HER2 status | 0.430 | ||
| Positive | 33 (37.1) | 28 (31.5) | |
| Negative | 56 (62.9) | 61 (68.5) | |
| Subsequent therapy | |||
| Chemotherapy | 59 (66.3) | 67 (75.3) | 0.187 |
| Endocrinotherapy | 58 (65.2) | 54 (60.7) | 0.535 |
| Targeted therapy | 33 (37.1) | 28 (31.5) | 0.430 |
Data were presented as mean ± standard deviation or count (percentage). Comparison was determined by t-test or Chi-square test. P<0.05 was considered as significant. LncRNA, long non-coding RNA; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
LncRNA SNHG1 expression in tumor tissue and its correlation with OS
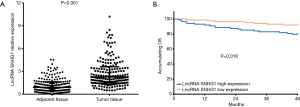
The expression of lncRNA SNHG1 in tumor tissue was elevated compared with the adjacent tissue (P<0.001, Figure 1A). K-M curves and log-rank test showed that lncRNA SNHG1 high expression patients exhibited shorter accumulating OS compared to lncRNA SNHG1 low expression patients (P=0.016, Figure 1B).
LncRNA SNHG1 expression among different cell lines
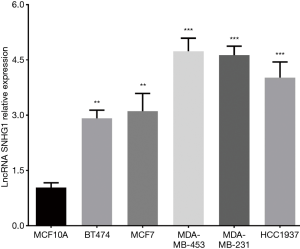
As presented in Figure 2, all five breast cancer cell lines including BT474 (P<0.01), MCF7 (P<0.01), MDA-MB-453 (P<0.001), MDA-MB-231 (P<0.001) and HCC1937 (P<0.001) cell lines presented with elevated lncRNA SNHG1 level compared to normal breast epithelial cell line MCF10A. Subsequently, MDA-MB-453 cell line and MCF7 cell line were chosen for further investigation.
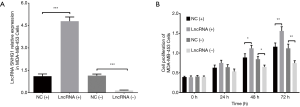
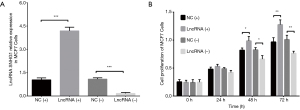
Cell proliferation in MDA-MB-453 and MCF7 cells after transfections of lncRNA SNHG1 (+/−) plasmids
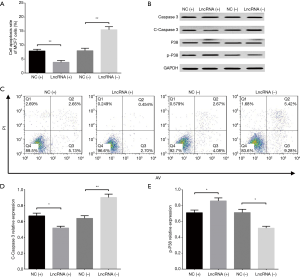
After transfection of lncRNA SNHG1 (+/−) plasmids into MDA-MB-453 cells, lncRNA SNHG1 expression was increased in lncRNA (+) group compared to NC (+) group (P<0.001) but decreased in lncRNA (−) group compared to NC (−) group (P<0.001), implying that the transfections were successful (Figure 3A). MDA-MB-453 cell proliferation after transfection was evaluated by CCK8 assay, which revealed that cell proliferation was enhanced in lncRNA (+) group compared to NC (+) group, while it was repressed in lncRNA (−) group compared to NC (−) group both at 48 h (both P<0.05) and 72 h (both P<0.01) after transfections (Figure 3B). LncRNA SNHG1 (+/−) plasmids were also transfected into MCF7 cells successfully (all P<0.001, Figure 4A), then similar results were observed in MCF7 cells consequently (all P<0.05, Figure 4B).
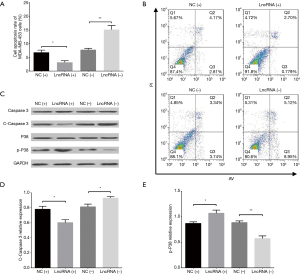
Cell apoptosis in MDA-MB-453 and MCF7 cells after transfections of lncRNA SNHG1 (+/−) plasmids
AV/PI assay revealed that cell apoptosis rate in MDA-MB-453 cells was decreased in lncRNA (+) group compared to NC (+) group (P<0.05) but increased in lncRNA (−) group compared to NC (−) group (P<0.01) at 72 h (Figure 5A,B). In addition, we also measured the expression of pro-apoptotic and anti-apoptotic markers in each group, which disclosed that compared to NC (+) group, lncRNA (+) group exhibited reduced C-Caspase 3 level (P<0.05, Figure 5C,D) but elevated p-P38 level (P<0.05, Figure 5C,E); meanwhile, lncRNA (−) group presented with increased C-Caspase 3 level (P<0.05, Figure 5C,D) while decreased p-P38 level compared to NC (−) group (P<0.01, Figure 5C,E). Similar results were also observed in MCF7 cells (all P<0.05, Figure 6A,B,C,D,E).
Discussion
In the current study we found that: (I) lncRNA SNHG1 expression in tumor tissue was up-regulated compared to adjacent non-tumor tissue, and it was positively correlated with T stage but negatively associated with OS in breast cancer patients. (II) LncRNA SNHG1 promoted breast cancer cell proliferation but suppressed cell apoptosis.
LncRNA SNHG1 is one of the most commonly investigated lncRNAs that act as pivotal modulators in human biological activities including gene expression and cell functions regulation. Since lncRNA SNHG1 is found to promote cell proliferation in non-small cell lung cancer (NSCLC) in 2014 for the first time, it has been reported to be closely associated with various cancers (17-19). An in vitro experiment reveals that lncRNA SNHG1 is upregulated in hepatocellular carcinoma (HCC) cell line (HepG2) compared to normal human liver cell line (L02), and it promotes HCC cell proliferation, invasion and migration via sponging microRNA (miR)-195 (20). Another in vitro study also illuminates that lncRNA SNHG1 expression is up-regulated in gastric cancer cell lines (NCI-N87 and MKN-45) and it enhances gastric cancer cell proliferation through regulating DNA (cytosine-5)-methyltransferase 1 (DNMT1) (21). These experiments suggest that lncRNA SNHG1 may be involved in the development and progression of cancers. More interestingly, several studies have revealed the negative correlation of lncRNA SNHG1 level with the prognosis of cancer patients (20,21). For example, a previous study elucidates that lncRNA SNHG1 expression in HCC tumor tissue is higher than that in adjacent normal tissue, and HCC patients with high lncRNA SNHG1 expression in tumor tissue exhibit shorter recurrence-free survival and OS compared to patients with low lncRNA SNHG1 expression (20). Meanwhile, the expression of lncRNA SNHG1 is also elevated in gastric cancer tissue compared to the matched adjacent non-tumor tissue, and the OS of gastric cancer patients with high expression of lncRNA SNHG1 in tumor tissue is lower than that of patients with low expression of lncRNA SNHG1 (21). However, the lncRNA SNHG1 expression level in breast cancer patients and its correlation with the prognosis of breast cancer patients is still unclear. Hence, we conducted the current study and discovered that lncRNA SNHG1 expression was increased in breast cancer tissue compared to adjacent non-tumor tissue, and the elevated lncRNA SNHG1 expression in tumor tissue was correlated with the worse OS. The possible explanation might be that: as previously described, lncRNA SNHG1 might promote the progression of breast cancer through enhancing cancer cell proliferation, migration and invasion while inhibiting cancer cell apoptosis via regulating targeting miRNAs or downstream genes such as p38, p53 (20-24). In the present study, we also observed that there was no difference in breast cancer molecular subtypes between lncRNA SNHG1 low expression patients and lncRNA SNHG1 high expression patients, which might be due to that lncRNA SNHG1 was involved in the pathogenesis of breast cancers with different molecular subtypes thereby contributed to poorer OS. However, the detailed association of lncRNA SNHG1 expression with molecular subtypes of breast cancer needs to be verified in further exploration.
The effect of lncRNA SNHG1 on cancer cell proliferation and apoptosis is discovered by previous studies in various cancers (25,26). Besides, lncRNA SNHG1 is also observed to promote cell proliferation, migration and invasion while reduce cell apoptosis in many other cancer cell lines (25-28). For instance, lncRNA SNHG1 enhances cell growth while reduces cell apoptosis in esophageal squamous cell carcinoma cells (obtained from esophageal squamous cell carcinoma patients) by directly binding to miR-338-3p (29). In the meantime, another experiment illuminates that lncRNA SNHG1 regulates expression of zinc finger E-box binding homeobox 1 (ZEB1) by interacting with p63 TA isoform (TAp63), and promotes cell proliferation, migration as well as invasion but inhibits cell apoptosis in lung squamous cell carcinoma (SCC) cell lines H-266 and SK-MES-1 (30). To testify the hypothesis that lncRNA SNHG1 might display similar functions in breast cancer cells, in vitro experiments were conducted in this study, which disclosed that lncRNA SNHG1 promoted cell proliferation while repressed cell apoptosis in breast cancer cells. This might be due to that: lncRNA SNHG1 might regulate multiple pathways to promote cell proliferation and repress cell apoptosis of breast cancer cells, for example, mediating its neighboring gene SLC3A2 and then affecting integrin-mediated focal adhesion kinase (FAK)/PI3K/AKT signaling pathway, or via modulating the activities of p53 and some miRNAs such as miR-145, miR-424-5p and miR-130a-3p (12,18,22,31,32).
In conclusion, lncRNA SNHG1 is upregulated in tumor tissue and correlates with higher T stage and worse OS, and it promotes cell proliferation but inhibits cell apoptosis in breast cancer.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Board of The Central Hospital of Wuhan (No. 2011-12), all the patients provided signed informed consents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin 2016;66:31-42. [Crossref] [PubMed]
- Li T, Mello-Thoms C, Brennan PC. Descriptive epidemiology of breast cancer in China: incidence, mortality, survival and prevalence. Breast Cancer Res Treat 2016;159:395-406. [Crossref] [PubMed]
- DeSantis CE, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67:439-48. [Crossref] [PubMed]
- Carroll JS, Hickey TE, Tarulli GA, et al. Deciphering the divergent roles of progestogens in breast cancer. Nat Rev Cancer 2017;17:54-64. [Crossref] [PubMed]
- Harbeck N, Gnant M. Breast cancer. Lancet 2017;389:1134-50. [Crossref] [PubMed]
- Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337-46. [Crossref] [PubMed]
- Mathieu EL, Belhocine M, Dao LT, et al. Functions of lncRNA in development and diseases. Med Sci (Paris) 2014;30:790-6. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Wu Z, Liu X, Liu L, et al. Regulation of lncRNA expression. Cell Mol Biol Lett 2014;19:561-75. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Taheri M, Omrani MD, Ghafouri-Fard S. Long non-coding RNA expression in bladder cancer. Biophys Rev 2018;10:1205-13. [Crossref] [PubMed]
- Liu Y, Yang Y, Li L, et al. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol 2018;96:38-43. [Crossref] [PubMed]
- Wang Q, Li Q, Zhou P, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother 2017;91:906-11. [Crossref] [PubMed]
- Yang H, Wang S, Kang YJ, et al. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol Rep 2018;40:261-71. [PubMed]
- You J, Fang N, Gu J, et al. Noncoding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian J Cancer 2014;51:e99-102. [Crossref] [PubMed]
- Sun Y, Wei G, Luo H, et al. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene 2017;36:6774-83. [Crossref] [PubMed]
- Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med 2019;23:2351-61. [Crossref] [PubMed]
- Zhang H, Zhou D, Ying M, et al. Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195. Med Sci Monit 2016;22:4820-9. [Crossref] [PubMed]
- Hu Y, Ma Z, He Y, et al. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun 2017;491:926-31. [Crossref] [PubMed]
- Shen Y, Liu S, Fan J, et al. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep 2017;18:536-48. [Crossref] [PubMed]
- Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun 2017;487:146-52. [Crossref] [PubMed]
- Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/beta-catenin pathway. Biochem Biophys Res Commun 2018;495:238-45. [Crossref] [PubMed]
- Lu Q, Shan S, Li Y, et al. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J 2018;32:3957-67. [Crossref] [PubMed]
- Wang J, Cao L, Wu J, et al. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol 2018;52:77-88. [PubMed]
- Sun X, Wang Z, Yuan W. Down-regulated long non-coding RNA SNHG1 inhibits tumor genesis of colorectal carcinoma. Cancer Biomark 2017;20:67-73. [Crossref] [PubMed]
- Zhang Y, Jin X, Wang Z, et al. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark 2017;21:89-96. [Crossref] [PubMed]
- Yan Y, Fan Q, Wang L, et al. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget 2017;8:35750-60. [PubMed]
- Zhang HY, Yang W, Zheng FS, et al. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed Pharmacother 2017;90:650-8. [Crossref] [PubMed]
- Kong X, Zhang J, Li J, et al. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem Biophys Res Commun 2018;501:486-93. [Crossref] [PubMed]
- Wang J, Wang S, Zhou J, et al. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother 2018;102:147-52. [Crossref] [PubMed]


