
GSTM1 polymorphism in Oncotype DX assay is a potential predictive factor for taxane-based neoadjuvant chemotherapy in estrogen receptor-positive Chinese breast cancer patients
Introduction
With the development and application of molecular biology, breast cancer can be classified into levels of malignant biological behavior via multiple gene expression detections into molecular subtypes. The 21-gene Oncotype Dx was reported as one of the multigene assays providing significant predictive power to breast cancer prognosis and grading the patients’ benefit from chemotherapy. The recurrence score has been demonstrated to be effective and accurate in ER-positive patients in various clinical studies (1-3). For neoadjuvant settings, however, the prediction value of RS for response has not been adequately investigated, and it is still unclear whether any individual gene of the 16 cancer related genes out of the 21-gene panel can be used to predict response to taxane-based neoadjuvant chemotherapy.
GSTM1 in Oncotype DX assay is one of the members in the glutathione-S-transferase (GST) family that can catalyze the conjugation of glutathione (GSH) to a wide variety of endogenous and exogenous electrophilic compounds for Phase II detoxification, including antineoplastic agents (4-6). Human GSTs are categorized into 6 classes (α, µ, π, σ, ω, θ, and ζ) (4). However, the role of GSTs in the protection of cells against taxane remains unclear despite various studies (5-10).
This study aimed to investigate whether GSTM1 expression levels could act as a predictive marker for responses to taxane-based chemotherapy in breast cancer patients. First, we measured GSTM1 expression levels from tumor tissues and tested their potential links to chemotherapy responses within a cohort of Chinese breast cancer patients. Second, we set up cell models to further explore the possible molecular mechanisms to accredit such correlations biologically. By such a two-step designed study, the hypothesis was adequately tested and the results were solid to suggest the predictive role of GSTM1.
Methods
Patients and therapy response
The study retrospectively included 66 Chinese patients diagnosed with stage I to III invasive breast cancer, who were submitted to the taxane-based neoadjuvant chemotherapies at the Department of Breast Cancer in Guangdong Provincial People’s Hospital, Guangzhou, China, between February 2012 and December 2014. Inclusion criteria were as follows: (I) a single, unilateral tumor without clinical or radiological signs of metastasis; (II) patient age between 35 and 70 years; (III) estrogen- and/or progesterone-receptor-positive (ER+/PgR+); and (IV) previously stored fresh frozen tumor tissues before treatment. All patients underwent 4 cycles of taxane-based regimens at 21-day intervals before surgery. For HER2-positive patients, trastuzumab was incorporated concurrently with chemotherapy. The clinical response to chemotherapy was evaluated according to RECIST criteria. Pathological complete response (pCR) was defined as no histological evidence of residual invasive cancer both in the breast and in the axilla.
Patient gene profiling in by qRT-PCR
Tumor tissues in 66 enrolled Chinese ER-positive breast cancer patients were subjected to TaqMan RT-PCR reactions, performed exactly as described previously for the assay for the 21-gene panel (3,11). Expression levels were normalized relative to the set of 5 reference genes. All reactions were repeated 3 times. The 16 cancer-related genes were Ki67, STK15, survivin, cyclin B1, MYBL2, GRB7, HER2, ER, PgR, BCL2, SCUBE2, MMP11, CTSL2, GSTM1, CD68, and BAG1, and the 5 reference genes were ACTB, GAPDH, PLPO, GUS, and TFRC.
Statistics analysis
The analysis consisted in comparing clinical response with isolated genes. Data were input into the Statistical Program for the Social Sciences (SPSS, version 13.0, SPSS Inc., Chicago, IL, USA) and analyzed for statistical significance using the univariate/multivariate logistic regression analysis as well as the chi-square test. Overall significance of each gene expression was determined by calculating the odds ratios (OR). The odds ratio was expressed with a corresponding 95% confidence interval (CI). P<0.05 was considered to be statistically significant.
Cell culture and experimental reagents
Human breast cancer cell lines MDA-MB-231, docetaxel (TXT)-resistant SKBR3, TXT-resistant BT474, TXT-resistant MCF7, and TXT-resistant MDA-MB-231 were obtained from Dr. Wang HH. All of these cell lines were cultured according to the ATCC instructions. siRNA and Lipofectamine™ 2000 were purchased from Thermo Scientific Dharmacon®. SYBR Green Real-time PCR Master Mix was purchased from Toyobo (Osaka, Japan). RevertAid™ First Strand cDNA Synthesis kit was obtained from MBI (Fermentas, Hanover, MD, USA). Docetaxel (Aventis Pharmaceuticals, Bridgewater, NJ, USA) was stored at a concentration of 10 mg/mL (12.6 mM) in 13% w/w ethanol at 4 °C and diluted in medium before use. Cell Counting Kit-8 was obtained from Roche. TUNEL assay was performed by use of In Situ Cell Death Detection Kit (Fluorescein, Roche, Switzerland). DAPI (4,6-diamidino-2-phenylindole) was purchased from Beyotime (Shanghai, China). Hoechst 33342 was purchased from Sigma-Aldrich China (Shanghai, China).
Cell fractionation and Western blot analysis
The expression of GST isozymes in 5 breast cancer cells was investigated as described previously (12). Cytosolic proteins (20 µg) were separated by 12% SDS-polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose membrane. The membranes were blocked overnight followed by incubation with the mouse monoclonal primary antibodies anti-GSTA1, M1, P1, or T1 1:500 (Sigma), and mouse monoclonal primary antibody anti-β-actin 1:2,000 (Sigma). Then, membranes were incubated with anti-mouse peroxidase-conjugated secondary antibodies 1:5,000 for GSTs and β-actin. Specific proteins were visualized using X-ray films after incubation with enhanced chemiluminescence reagents (Amersham Biosciences, Corston, UK).
Real-time RT-PCR
Extraction of total cellular RNA was carried out using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol, and expression was analyzed by RT-PCR using standard procedures as described previously (13). The primer sequences for β-actin, GSTA1, GSTP1, GSTM1, GSTT1, which were purchased from Santa Cruz, are available on request. PCR products were run on agarose gels supplemented with ethidium bromide and visualized by ultraviolet illumination. Band intensities were quantified by densitometry analysis using Bio1D software (Vilber Lourmat, Marne La Vallée, France).
Knockdown of GSTA1, GSTP1, GSM1, and GSTT1 with siRNA oligos
GSTA1si, GSTP1si, GSTM1si, and GSTT1si stock solutions (20 µM) were diluted with 1× siRNA buffer to form 5 µM solutions. Lipofectamine RNAiMAX transfection reagent was mixed with 5 µM siGST incubated for 20 min. The final concentration of siRNA was 50 nM. The cells were incubated with siRNA for 24 and 48 h for subsequent experiments.
Measurement of docetaxel sensitivity
Cells (1×104) were seeded into a flat bottomed 96-well plate and incubated with various concentrations of docetaxel for 24 h at 37 °C. Cell viability was determined by the -(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)(2H)tetrazolium monosodium salt (WST) assay using Cell Counting Kit-8 according to the manufacturer’s instructions, and the percentage of cell death was calculated as described previously (14). To measure docetaxel sensitivity after GST isozyme knockdown, docetaxel was added to the culture medium at 24 h post-transfection with siRNA treatment.
Hoechst staining
Following treatment with 100 µM docetaxel for 24 h, the MDA-MB-231 cells were harvested by centrifugation at 800 rpm for 5 min, washed with PBS, and fixed with methanol acetic acid for 10 min. Fixed cells were washed with PBS followed by staining with 200 µM Hoechst 3325 (1 mg/mL) at room temperature in the dark for 5 min. Hoechst 33258 stained cells were examined and immediately photographed with a fluorescence microscope with an excitation wavelength of 330–380 nm.
TUNEL assay
TUNEL assay was performed as previously described (15). Briefly, following treatment with 100 µM docetaxel for 24 h, the MDA-MB-231 cells were fixed with 4% paraformaldehyde in PBS for 10 min, washed 3 times with PBS, and then stained with TUNEL following manufacturer’s instructions or with DAPI (0.5 µg/mL) for 10 min at room temperature. Images of the cells were then acquired using fluorescence microscope.
Results
GSTM1 mRNA expression correlates with taxane-neoadjuvant therapy response
The 66 patients, with an average age 51.7 years, were ER positive by IHC, and 36.4% (n=24) were HER2 positive. Twenty-five patients achieved pCR, 14 of whom were HER2-positive patients, and 11 of whom were HER2-negative patients (58.3% vs. 26.2%, chi-square value =6.706, odds ratio =3.945, 95% CI: 1.362–11.431, P=0.01). Analysis of the results of patients is summarized in Table 1. It was noted that only GSTM1 has distinct non-expression/expression polymorphism, with 33.3% (n=22) non-expression vs. 66.7% expression; this polymorphism does not correlate to any other assayed genes in this study. Univariate analysis showed that the expression level of HER2 and GSTM1 non-expression (rather than expression level) correlate to pCR at the significance level 0.05 (GSTM1: odds ratio 0.349 and P=0.048; HER2: odds ratio 3.945 and P=0.010; GRB7: odds ratio 1.0 and P=0.043; cyclin B: odds ratio 1.0 and P=0.024). We further included HER2 and GSTM1 in multivariate logistic regression analysis. However, none of these 2 genes showed statistical significance for pCR.
Table 1
| Genotype | pCR (n=25) | Non-pCR (n=41) | Odds ratio (95% CI) | P |
|---|---|---|---|---|
| GSTM1 | 0.349 (0.121–1.009) | 0.048 | ||
| Expression | 13 | 31 | ||
| No-expression | 12 | 10 | ||
| HER2 | 3.945 (1.362–11.431) | 0.01 | ||
| Positive | 14 | 10 | ||
| Negative | 11 | 31 |
Differential expression of GST isoenzymes
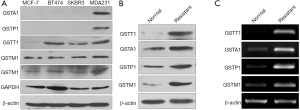
The expression of GSTM1, GSTA1, GSTP1, and GSTT1 varied across the 5 breast cancer cell lines (Figure 1A). Our results showed TXT-resistant MDA-MB-231 expresses all the 4 GST isozymes at higher levels compared to other lines. TXT-resistant MCF7 cells express GSTM1 at lower levels; GSTT1 levels are barely detectable whereas GSTA1 and GSTP1 were not detected. SKBR3 and BT474, the HER2-positive cells, express GSTM1 at lower levels compared to TXT-resistant MDA-MB-231 and also express GSTT1. Interestingly, our results showed a significant increase in GSTM1, GSTP1, GSTA1, and GSTT1 expression, measured at mRNA levels and protein levels (Figure 1B,C), in TXT-resistant MDA-MB-231 cells compared to non-resistant MDA-MB-231 cells.
Docetaxel sensitivity in human breast cancer cell lines with differential expression of GST isozymes
We next tested if variation in sensitivity to docetaxel of human breast cancer cells could be correlated with expression of specific GST isoenzymes utilizing CCK8 assay for the analysis of cytotoxicity (Figure 2). Our results revealed a wide range of sensitivity among the 4 TXT-resistant cell lines for docetaxel IC50 values. Interestingly, taxane-resistant MDA-MB-231, with the highest expression of 4 GST isozymes, showed the highest IC50 for docetaxel (~7.3 µM). Resistant MCF7, which expresses relatively lower levels of GSTM1, was more sensitive to docetaxel (IC50: ~3.8 µM). Cell lines showing relatively more resistance to docetaxel (SKBR3 and BT474) were associated with higher GSTM1 levels and GSTT1 than the most sensitive cell lines (MCF-7). Moreover, normal MDA-MB-231 (IC50: ~33.0 nM) cells express significantly lower levels GST isozymes GSTM1, GSTP1, GSTA1, and GSTT1 (Figure 1B,C) compared to docetaxel resistant MDA-MB-231. Taken together, the breast cancer tumor cells expressing higher levels of GST isoenzyme(s) were more resistant to docetaxel, suggesting the levels of the GST isoenzymes correlate with sensitivity to docetaxel.

Depletion of GST isozymes in taxane-resistant MDA-MB-231 cells
Since our results showed taxane-resistant MDA-MB-231 cells express all 4 GST isozymes, we utilized these cells to study the individual contribution of each isozyme to docetaxel resistance. We used 2 different siRNA oligos targeting each of the 4 GST isozymes in order to achieve specific knockdown of their mRNA. By analyzing both protein lysates with enzyme specific antibodies in western blotting and mRNA message quantified by RT-PCR, we found a significant depletion of 4 isozymes 24 h post siRNA treatment (Figure 3). For docetaxel resistance assays, siRNA oligos that achieved maximum knockdown (si-GSTA1 02, si-GSTP1 02, si-GSTM1 01, and si-GSTT1 02) of each isozyme in resistant-MDA-MB-231 were selected.
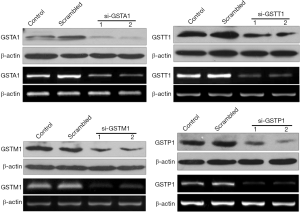
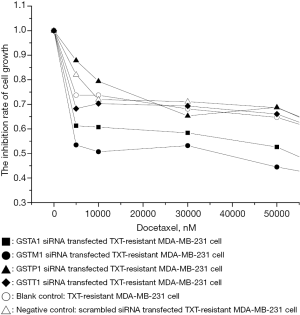
GSTM1 and GSTA1 depletion increased docetaxel sensitivity of taxane-resistant MDA-MB-231 cells
Taxane-resistant MDA-MB-231 were most resistant to docetaxel which, interestingly expressed higher levels of all 4 GST isozymes compared with normal MDA-MB-231 cells (Figure 3). In order to identify the role of each isozyme in docetaxel resistance, we analyzed the sensitivity to docetaxel using WST assay over a period of 24 h (Figure 4) after knocking down each isozyme (Figure 3) in MDA-MB-231cells. Our results showed that the depletion of GSTA1 (oligo: si-GSTA1 02) enhanced the docetaxel sensitivity by 138% in resistant MDA-MB-231 cells compared with scrambled siRNA-treated or cells with no siRNA treatment (i.e., decrease in IC50 value by 0.5-fold, from ~7.3 µM to ~4.7 µM). GSTM1 knockdown (oligo: si-GSTM1 02) resulted in a docetaxel IC50 of ~4.2 µM compared to a control cell IC50 of ~2 M. In notable contrast, GSTP1 and GSTT1 depletion had no effect on docetaxel sensitivity compared to controls (IC50 ~0.9 M for both). Collectively, the results demonstrate that GSTA1 and GSTM1 levels, but not GSTP1 or GSTT1 levels, are inversely correlated with docetaxel-induced cell cytotoxicity.
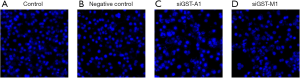
Evaluation of apoptosis by Hoechst staining in GSTA1 and GSTM1-depleted resistant-MDA-MB-231 cells
Apoptosis is one of the principal mechanisms of docetaxel induced cytotoxicity. We evaluated the apoptotic morphology in GSTA1- and GSTM1-depleted resistant-MDA-MB-231 cells after treatment with docetaxel (100 µM) for 24 h using membrane-permeable blue Hoechst 33258 (Figure 5). Our results showed nuclei of docetaxel-treated GSTA1- and GSTM1-depleted cells appeared hypercondensed (brightly stained). The number of apoptotic nuclei containing nuclear fragmentation and condensed chromatin induced by docetaxel increased significantly as the result of GSTA1 and GSTM1 knockdown. Similar treatment in control cultures showed nuclei with regular contours that were round and large in size. Cells with smaller nuclei and condensed chromatin were rarely seen. Scrambled siRNA treatment did not change the nuclear morphology of docetaxel-treated cells.
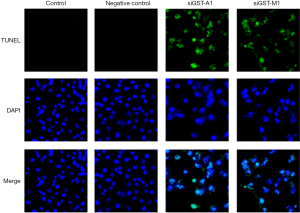
Evaluation of apoptosis by TUNEL staining in GSTA1- and GSTM1-depleted resistant-MDA-MB-231 cells
We next used TUNEL assay for quantification of the docetaxel-induced apoptosis in GSTM1 and GSTA1 knock-down cells. Consistent with Hoechst staining results, TUNEL analysis of GSTM1 and GSTA1 knock-down cells after treatment with docetaxel (100 µM) for 24 h resulted in significant TUNEL-positive nuclei (green spots) resulting from staining of nicked DNA by FL-dUTP (Figure 6). DAPI staining of GSTM1 and GSTA1 knock-down cells showed fragmented nuclei and enhanced DAPI fluorescence by densely stained nuclear granular bodies (apoptotic bodies). Control cells (blank and scramble siRNA) displayed a homogenous morphology with nuclei lightly and evenly stained by DAPI (Figure 6).
Discussion
In 66 ER-positive Chinese breast cancer patients, GSTM1 had a distinct loss or variation in expression, which was significantly associated with increased response rate to taxane-based neoadjuvant chemotherapy. Our findings are consistent with a retrospective study by Ambrosone et al. that showed greater recurrence-free survival among patients with GSTM1-null genotypes than patients with wild GSTM1 (16). Similarly, the observations of Khedhaier et al. in 309 breast cancer patients are consistent with our results (17). However, studies by Lizard-Nacol et al. and Yang et al. showed no association of treatment response among patients with the wild type or polymorphic GSTM1 in women treated with neoadjuvant or adjuvant chemotherapy (18,19). Although the observed association between GSTM1 polymorphisms and neoadjuvant outcomes in our study did reach significance, a multivariate logistic regression analysis of HER2 and GSTM1 for pCR did not show statistical significance, most probably due to the small number of patients enrolled in this study to guarantee statistical validity.
Our results from 5 breast cancer cell lines showed an inverse correlation with GST isoenzymes expression level and sensitivity to docetaxel. This observation is in agreement with recent findings by Armstrong et al. for 6 human breast cancer cell lines (20). We then used TXT-resistant MDA-MB-231 cells that expressed all 4 GST isozymes to study the functional role of each isozyme in docetaxel resistance by siRNA. Our data showed a 1.5-fold increase in docetaxel sensitivity upon GSTA1 and GSTM1 knockdown in comparison to controls, and observed significantly enhanced docetaxel-induced apoptosis, suggesting the role of GSTM1 and GSTA1 as the resistance factors responsible for intrinsic resistance to docetaxel. In contrast, GSTP1 and GSTT1 knockdown resulted in no significant change to docetaxel sensitivity.
In summary, our findings strongly supported the current understanding of GSTM1 involvement in docetaxel detoxification and for the first time demonstrated that inhibition of GSTM1 and GSTA1 contributes to overcoming docetaxel resistance in breast cancer lines. GSTM1 expression was identified as a predictive marker in patients with breast cancer that underwent taxane-based neoadjuvant chemotherapy.
Acknowledgments
Funding: This study has been supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was waived due to the retrospective nature of the study. Institutional Review Board approval was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cronin M, Sangli C, Liu ML, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem 2007;53:1084-91. [Crossref] [PubMed]
- Mamounas EP, Tang G, Fisher B, et al. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol 2010;28:1677-83. [Crossref] [PubMed]
- Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817-26. [Crossref] [PubMed]
- Sau A, Pellizzari Tregno F, Valentino F, et al. Glutathione transferases and development of new principles to overcome drug resistance. Arch Biochem Biophys 2010;500:116-22. [Crossref] [PubMed]
- Arai T, Miyoshi Y, Kim SJ, et al. Association of GSTP1 expression with resistance to docetaxel and paclitaxel in human breast cancers. Eur J Surg Oncol 2008;34:734-8. [Crossref] [PubMed]
- Iwao-Koizumi K, Matoba R, Ueno N, et al. Prediction of docetaxel response in human breast cancer by gene expression profiling. J Clin Oncol 2005;23:422-31. [Crossref] [PubMed]
- Park JS, Yamamoto W, Sekikawa T, et al. Cellular sensitivity determinants to docetaxel in human gastrointestinal cancers. Int J Oncol 2002;20:333-8. [PubMed]
- Romero A, Martin M, Oliva B, et al. Glutathione S-transferase P1 c.313A > G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Ann Oncol 2012;23:1750-6. [Crossref] [PubMed]
- Schmidt M, Bachhuber A, Victor A, et al. p53 expression and resistance against paclitaxel in patients with metastatic breast cancer. J Cancer Res Clin Oncol 2003;129:295-302. [PubMed]
- Wang L, Jiang Z, Sui M, et al. The potential biomarkers in predicting pathologic response of breast cancer to three different chemotherapy regimens: a case control study. BMC Cancer 2009;9:226. [Crossref] [PubMed]
- Cronin M, Pho M, Dutta D, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol 2004;164:35-42. [Crossref] [PubMed]
- Wang CH, Wu HT, Cheng HM, et al. Inhibition of glutathione S-transferase M1 by new gabosine analogues is essential for overcoming cisplatin resistance in lung cancer cells. J Med Chem 2011;54:8574-81. [Crossref] [PubMed]
- Depeille P, Cuq P, Mary S, et al. Glutathione S-transferase M1 and multidrug resistance protein 1 act in synergy to protect melanoma cells from vincristine effects. Mol Pharmacol 2004;65:897-905. [Crossref] [PubMed]
- Kojima Y, Nakayama M, Nishina T, et al. Importin beta1 protein-mediated nuclear localization of death receptor 5 (DR5) limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced cell death of human tumor cells. J Biol Chem 2011;286:43383-93. [Crossref] [PubMed]
- Lopez-Pulido EI, Munoz-Valle JF, Del Toro-Arreola S, et al. High expression of prolactin receptor is associated with cell survival in cervical cancer cells. Cancer Cell Int 2013;13:103. [Crossref] [PubMed]
- Ambrosone CB, Sweeney C, Coles BF, et al. Polymorphisms in glutathione S-transferases (GSTM1 and GSTT1) and survival after treatment for breast cancer. Cancer Res 2001;61:7130-5. [PubMed]
- Khedhaier A, Remadi S, Corbex M, et al. Glutathione S-transferases (GSTT1 and GSTM1) gene deletions in Tunisians: susceptibility and prognostic implications in breast carcinoma. Br J Cancer 2003;89:1502-7. [Crossref] [PubMed]
- Lizard-Nacol S, Coudert B, Colosetti P, et al. Glutathione S-transferase M1 null genotype: lack of association with tumour characteristics and survival in advanced breast cancer. Breast Cancer Res 1999;1:81-7. [Crossref] [PubMed]
- Yang G, Shu XO, Ruan ZX, et al. Genetic polymorphisms in glutathione-S-transferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Cancer 2005;103:52-8. [Crossref] [PubMed]
- Armstrong DK, Gordon GB, Hilton J, et al. Hepsulfam sensitivity in human breast cancer cell lines: the role of glutathione and glutathione S-transferase in resistance. Cancer Res 1992;52:1416-21. [PubMed]


