Correlative analysis of gene mutation and clinical features in patients with non-small cell lung cancer
Introduction
Lung cancer is the main reason for death associated with cancer death all over the world (1). With the increase of morbidity and mortality, cancer has become the main cause of death and an important public health problem in China where cancer is the most common cancer (2). Nearly 733,300 new cases and 610,200 deaths of lung cancer were estimated to have occurred in China in 2015 (2).
More than 85% of lung cancer cases are non-small cell lung cancer (NSCLC) (3). In the 90s, patients with advanced NSCLC were usually treated with chemotherapeutic drugs, particularly platinum drugs, regardless of histological subtype (4). The criteria for treatment of NSCLC began to change at the beginning of this century. The discovery of epidermal growth factor receptor (EGFR) mutations in NSCLC laid the foundation for the development of targeted molecular therapies and predictive biomarkers (5-8). For instance, in advanced NSCLC patients with EGFR mutation-positive disease in Asia, afatinib significantly improves progression-free survival with tolerable and controlled safety (9). Also, Wang et al. found that erlotinib monotherapy is more cost-effective than platinum-based chemotherapy as first-line treatment of EGFR mutation-positive NSCLC patients in the Chinese health care system (10).
Currently, more and more mutations are being found driving the mechanisms of NSCLC, especially in lung adenocarcinoma (ADC). Common mutations in the EGFR include KRAS, HER2, PIK3CA, BRAF, and MET genes, while gene rearrangements in ALK, ROS1, and RET are also common. Molecular-targeted drug therapy has been established as superior to chemotherapy in the treatment of gene mutations in NSCLC (6,11-14). Seventy percent of Chinese patients with ADC in the study harbored one of the four gene mutations (EGFR, KRAS, MET, and ALK mutations) (15). Therefore, for the diagnosis and treatment of NSCLC, the NCCN guidelines have recommended testing for the gene mutations of KRAS, ALK, ROS1, EGFR, ERBB2, BRAF, RET, and MET (16).
Previous studies have suggested that most cases of NSCLC patients only express one type of gene mutation, each gene mutation population has different clinicopathological features, and that each is expressed differently in different regions of the population (17). Sequist et al. found that 51% of the 552 smoking Caucasians diagnosed with NSCLC had a gene mutation, with the most common being KRAS (24%), EGFR (13%), PIK3CA (4%), and ALK (5%) (18). Additionally, Sun et al.’s study found that 90% of 52 non-smoking Chinese lung ADC patients had EGFR, KRAS, ALK, or HER2 gene mutations (19). There are also differences in the types of EGFR gene mutations in different regions. For example, in China, the EGFR gene mutation in patients in Kunming is mainly exon 19, while in Shanghai, the EGFR gene mutation is mainly in exon 19 and exon 21 (20).
At present, there are few studies on multiple driver genes and clinicopathological features of the population in Hunan, China. In this study, we aimed to analyze multiple mutation genes and clinical features of NSCLC patients in this region in order to learn its relevance.
Methods
Study subjects
The cases of this study were collected from Xiangya Hospital of Central South University. It is a large, comprehensive, tertiary first-class hospital with multiple departments including those in geriatrics, respiratory medicine, thoracic surgery, oncology, pathology, etc.
The inclusion criteria for subjects were as follows: (I) aged 18 or above; (II) pathologically confirmed as NSCLC, using bronchoscopy, thoracoscopy, biopsy, surgical resection or pleural effusion; (III) underwent next-generation sequencing (NGS) to complete the detection of eight genes (EGFR/ALK/ROS1/MET/ERBB2/RET/BRAF/KRAS); (IV) had complete medical records.
From February 2016 to December 2017, the Department of Geriatric Respiratory Medicine of Xiangya Hospital of Central South University diagnosed 113 cases of NSCLC, for which NGS genetic testing was performed, conforming to the inclusion criteria. Tumor staging was based on the 8th edition of TNM classification.
Clinical, pathological feature collection
Patient demographics and clinical characteristics including age, gender, smoking history, clinical diagnosis, pathology, TNM staging, and gene mutation information were collected from the database system of Xiangya Hospital of Central South University.
DNA preparation for NGS
According to the manufacturer’s instructions, tissue DNA was extracted and circulating cell-free DNA was recovered by using the QIAamp DNA FFPE tissue kit and the QIAamp Circulating Nucleic Acid kit (Qiagen, Valencia, CA, USA) respectively. DNA quantification was conducted using the Qubit 2.0 fluorimeter (Thermo Fisher Scientific, Waltham, MA, USA).
NGS library preparation
DNA shearing was conducted by using the Covaris M220 Focused Ultrasonicator (Covaris, Inc., Woburn, MA, USA), with subsequent end repair, phosphorylation, and adaptor ligation. Sized 200–400 bp fragments were selected by using the Agencourt AMPure XP Kit (Beckman Coulter, Fullerton, CA, USA), then hybridized with capture probes baits, selected with magnetic beads, and amplified with PCR. Indexed samples were sequenced with paired-end reads by using a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA). All sample genetic profiles were assessed by capture-based targeted deep sequencing using the 8-gene panel that contained the oncogenic driver mutations of KRAS, ALK, ROS1, EGFR, ERBB2, BRAF, RET, and MET.
Sequence data analysis
The sequence data analysis was performed as described previously (21,22). Sequence data were mapped to the human genome (hg19) by using Burrows-Wheeler Aligner. Local alignment optimization, mark duplication, and variant calling were conducted by using Genome Analysis Tool Kit, Picard, and VarScan. Gene rearrangements were called with Fusion And Chromosomal Translocation Enumeration and Recovery Algorithm, and copy number variations were analyzed according to sequencing depth. Variants were filtered by using the VarScan FP filter pipeline, with loci with depth less than 100 being filtered out. In both plasma and tissue samples, at least 5 supporting reads were needed for insertions and deletions, and 8 supporting reads were needed for single nucleotide variants. Variants with a population frequency over 0.1% were grouped as single nucleotide polymorphisms that were excluded from further analysis, while the remaining variants were annotated with SnpEff v3.6 and ANNOVAR.
Statistical analysis
Our data were analyzed by GraphPad Prism 6.01 statistical software. Chi-square test and Fisher’s test were used to compare the proportions. When the two-tailed P value was less than 0.05, the difference was considered significant.
Results
Clinical features in patients with NSCLC
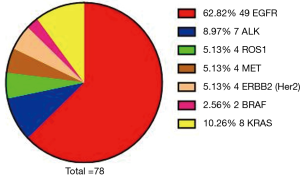
In this study, we included a total of 113 NSCLC cases, composed of 78 males and 35 females. In addition, there were 55 cases aged 60 or above and 58 cases aged lower than 60. According to the 8th edition of TNM classification, there were 95 patients of stage IV and 18 patients of non-stage IV. In the 113 cases, 67 of these people had a history of smoking. Histological distribution was characterized by ADC (78.76%) and squamous cell carcinoma (SCC, 21.24%). We found 71 cases of gene mutations. There was one concurrent mutation of ALK and ROS1, one concurrent mutation of EGFR and BRAF, one concurrent mutation of EGFR and MET, one concurrent mutation of MET and BRAF, and one concurrent mutation of EGFR and KRAS; there were two cases for concurrent mutation of EGFR and ERBB2. The distribution of each of the mutated genes was as follows: EGFR, 62.82%; ALK, 8.97%; ROS1, 5.13%; MET, 5.13%; ERBB2, 5.13%; RET, 0.00%; BRAF, 2.56%; KRAS, 10.26% (Table 1, Figure 1).
Table 1
| Clinicopathological features | Classification | Number of cases | Rate (%) |
|---|---|---|---|
| Gender | Male | 78 | 69.03 |
| Female | 35 | 30.97 | |
| Age | ≥60 | 55 | 48.67 |
| <60 | 58 | 51.33 | |
| TNM staging | IV | 95 | 84.07 |
| Non-IV | 18 | 15.93 | |
| Smoking history | Yes | 67 | 59.29 |
| No | 46 | 40.71 | |
| Pathological type | ADC | 89 | 78.76 |
| SCC | 24 | 21.24 | |
| Mutant genes | EGFR | 49 | 62.82 |
| ALK | 7 | 8.97 | |
| ROS1 | 4 | 5.13 | |
| MET | 4 | 5.13 | |
| ERBB2 (Her2) | 4 | 5.13 | |
| RET | 0 | 0.00 | |
| BRAF | 2 | 2.56 | |
| KRAS | 8 | 10.26 |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Gender distribution of mutated NSCLC patients and gender differences in patients with mutated NSCLC
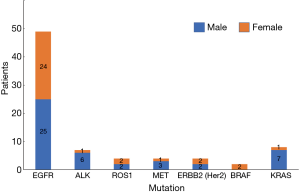
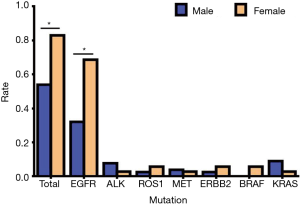
In our study, we found that 78 cases had genetic mutations, which included 45 males and 33 females. Among the patients with EGFR mutations, there were 25 males (51.02%) and 24 females (48.98%), in patients with ALK mutations there were 6 males (85.71%) and 1 female (14.29%), in patients with ROS1 mutations there were 2 males (50.00%) and 2 females (50.00%), in patients with MET mutations there were 3 males (75.00%) and 1 female (25.00%), in patients with ERBB2 (Her2) mutations there were 2 males (50.00%) and 2 females (50.00%), in patients with BRAF mutations there were 2 females (100.00%), and in patients with KRAS mutations there were 7 males (87.50%) and 1 female (12.50%) (Table 2, Figure 2). Our study showed that the total mutation rate was 53.85% in men and 82.86% in women (P<0.01), demonstrating that women are more susceptible to genetic mutations in NSCLC patients. Among the patients with EGFR mutations, the mutation rate was 32.05% in males and 68.57% in females (P<0.01), which proved that the female EGFR mutation rate was significantly higher than that of males, and there was a statistically significant difference. In the rest of the gene mutations, there was no significant difference in gender (P>0.05) (Table 3, Figure 3).
Table 2
| Mutated genes | Male | Female | |||
|---|---|---|---|---|---|
| Cases | Rate (%) | Cases | Rate (%) | ||
| EGFR | 25 | 51.02 | 24 | 48.98 | |
| ALK | 6 | 85.71 | 1 | 14.29 | |
| ROS1 | 2 | 50.00 | 2 | 50.00 | |
| MET | 3 | 75.00 | 1 | 25.00 | |
| ERBB2 (Her2) | 2 | 50.00 | 2 | 50.00 | |
| BRAF | 0 | 0.00 | 2 | 100.00 | |
| KRAS | 7 | 87.50 | 1 | 12.50 | |
Table 3
| Mutated genes | Male | Female | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Total | 42 | 53.85 | 29 | 82.86 | 0.0061 | |
| EGFR | 25 | 32.05 | 24 | 68.57 | 0.0003 | |
| ALK | 6 | 7.69 | 1 | 2.86 | 0.4330 | |
| ROS1 | 2 | 2.60 | 2 | 5.71 | 0.5865 | |
| MET | 3 | 3.85 | 1 | 2.86 | 1.0000 | |
| ERBB2 (Her2) | 2 | 2.56 | 2 | 5.71 | 0.5865 | |
| BRAF | 0 | 0 | 2 | 5.71 | 0.0940 | |
| KRAS | 7 | 8.97 | 1 | 2.86 | 0.4313 | |
Age distribution in of mutated NSCLC Patients and age differences in patients with mutated NSCLC
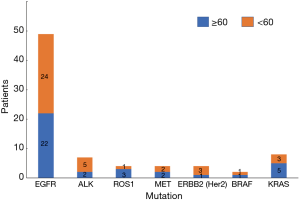
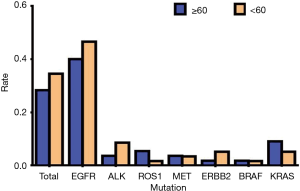
Of the 113 total NSCLC cases, 78 people had genetic mutations, with 36 people aged 60 or above and 42 people aged lower than 60. Among patients with EGFR mutations, there were 22 people aged 60 or above (44.90%) and 27 people less than 60 (55.10%); among the patients with ALK mutations, there were 2 people aged 60 or above (28.57%) and 5 patients aged lower than 60 (71.43%); among patients with ROS1 mutations, there were 3 patients aged 60 or above (75.00%) and 1 patient aged lower than 60 (25.00%); among patients with MET mutations, there were 2 patients aged 60 or above (50.00%) and 2 patients aged lower than 60 (50.00%); among patients with ERBB2 (Her2) mutations, there was 1 patient aged 60 or above (25.00%) and 1 patient aged lower than 60 (75.00%); among patients with BRAF mutations, there was 1 patient aged 60 or above (50.00%) and 1 patient aged lower than 60 (50.00%); among patients with KRAS mutations, there were 5 patients aged 60 or above (62.50%) and 3 patients aged lower than 60 (37.50%) (Table 4, Figure 4). Our data showed that the total mutation rate was 28.32% in patients aged 60 years and over, and the total mutation rate in patients under 60 years old was 34.51% (P>0.05), suggesting that there was no significant correlation between the mutations detected and age. In addition, there was no significant correlation with age in EGFR, ALK, and ROS1, etc., mutations (P>0.05) (Table 5, Figure 5).
Table 4
| Mutated genes | ≥60 years | <60 years | |||
|---|---|---|---|---|---|
| Cases | Rate (%) | Cases | Rate (%) | ||
| EGFR | 22 | 44.90 | 27 | 55.10 | |
| ALK | 2 | 28.57 | 5 | 71.43 | |
| ROS1 | 3 | 75.00 | 1 | 25.00 | |
| MET | 2 | 50.00 | 2 | 50.00 | |
| ERBB2 (Her2) | 1 | 25.00 | 3 | 75.00 | |
| BRAF | 1 | 50.00 | 1 | 50.00 | |
| KRAS | 5 | 62.50 | 3 | 37.50 | |
Table 5
| Mutated genes | ≥60 years | <60 years | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Total | 32 | 28.32 | 39 | 34.51 | 0.3192 | |
| EGFR | 22 | 40.00 | 27 | 46.55 | 0.4824 | |
| ALK | 2 | 3.64 | 5 | 8.62 | 0.4393 | |
| ROS1 | 3 | 5.45 | 1 | 1.72 | 0.3552 | |
| MET | 2 | 3.64 | 2 | 3.45 | 1.0000 | |
| ERBB2 (Her2) | 1 | 1.82 | 3 | 5.17 | 0.6188 | |
| BRAF | 1 | 1.82 | 1 | 1.72 | 1.0000 | |
| KRAS | 5 | 9.09 | 3 | 5.17 | 0.4823 | |
Pathological type distribution of mutated NSCLC Patients and pathological type differences in patients with mutated NSCLC
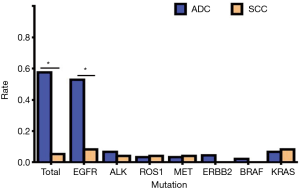
Of a total of 113 NSCLC cases, 78 people had genetic mutations, including 71 cases of ADC and 7 cases of SCC. In patients with EGFR mutations, there were 47 cases of ADC (95.92%) and 2 cases of SCC (4.08%); in patients with ALK mutations, there were 6 cases of ADC (85.71%) and 1 case of SCC (14.29%); in patients with ROS1 mutations, there were 3 cases of ADC (75.00%) and 1 case of SCC (25.00%); in patients with MET mutations, there were 3 cases of ADC (75.00%) and 1 case of SCC (25.00%); in patients with ERBB2 (Her2) mutations, there were 4 cases of ADC (100.00%); in patients with BRAF mutations, there were 2 cases of ADC (100.00%); in patients with KRAS mutations, there were 6 cases of ADC (75.00%) and 2 cases of SCC (25.00%) (Table 6, Figure 6). Our results showed that the total mutation rate of lung ADC was 57.52%, and the total mutation rate of lung SCC was 5.31% (P<0.01). The mutation rate of lung ADC proved to be significantly higher than in lung SCC. In patients with EGFR mutation, the mutation rate of ADC was 52.81%, the mutation rate of SCC was 8.33% (P<0.01). The mutation rate of EGFR in patients with lung ADC proved to be significantly higher than that found in patients with lung SCC. In other gene mutations, there was no significant difference in pathological types (P>0.05) (Table 7, Figure 7).
Table 6
| Mutated genes | ADC | SCC | |||
|---|---|---|---|---|---|
| Cases | Rate (%) | Cases | Rate (%) | ||
| EGFR | 47 | 95.92 | 2 | 4.08 | |
| ALK | 6 | 85.71 | 1 | 14.29 | |
| ROS1 | 3 | 75.00 | 1 | 25.00 | |
| MET | 3 | 75.00 | 1 | 25.00 | |
| ERBB2 (Her2) | 4 | 100.00 | 0 | 0 | |
| BRAF | 2 | 100.00 | 0 | 0 | |
| KRAS | 6 | 75.00 | 2 | 25.00 | |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
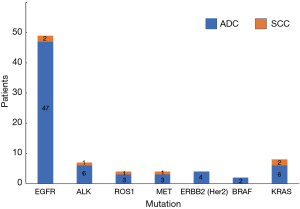
Table 7
| Mutated genes | ADC | SCC | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Total | 65 | 57.52 | 6 | 5.31 | <0.0001 | |
| EGFR | 47 | 52.81 | 2 | 8.33 | <0.0001 | |
| ALK | 6 | 6.74 | 1 | 4.17 | 1.0000 | |
| ROS1 | 3 | 3.37 | 1 | 4.17 | 1.0000 | |
| MET | 3 | 3.37 | 1 | 4.17 | 1.0000 | |
| ERBB2 (Her2) | 4 | 4.49 | 0 | 0 | 0.5767 | |
| BRAF | 2 | 2.25 | 0 | 0 | 1.0000 | |
| KRAS | 6 | 6.74 | 2 | 8.33 | 0.6768 | |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Smoking status of mutated NSCLC Patients and smoking status differences in patients with mutated NSCLC
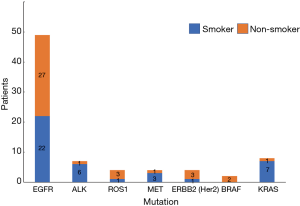
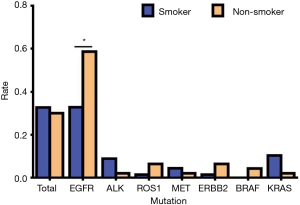
Of a total of 113 NSCLC cases, 78 people had genetic mutations including 40 smokers and 38 non-smokers. Among patients with EGFR mutations, there were 22 smokers (44.90%) and 27 non-smokers (55.10%); in patients with ALK mutations, there were 6 smokers (85.71%) and 1 non-smoker (14.29%); in patients with ROS1 mutations, there was 1 smoker (25.00%) and 3 non-smokers (75.00%); in patients with MET mutations, there were 3 smokers (75.00%) and 1 non-smoker (25.00%); in patients with ERBB2 (Her2) mutations, there was 1 smoker (25.00%) and 3 non-smokers (75.00%); in patients with BRAF mutations, there were 2 non-smokers (100.00%); in patients with KRAS mutations, there were 7 smokers (87.50%) and 1 non-smoker (12.50%) (Table 8, Figure 8). Our data analysis showed that the total mutation rate was 32.74% in smokers and 30.09% in non-smokers (P>0.05), indicating that the total incidence of gene mutations in NSCLC patients we detected was not significantly associated with smoking. However, among the patients with EGFR mutations, the mutation rate was 32.84% in smokers and 58.70% in non-smokers (P<0.01), demonstrating that non-smokers have significantly higher EGFR mutation rates than smokers. Among the other gene mutations, there was no significant difference between smokers and non-smokers (P>0.05) (Table 9, Figure 9).
Table 8
| Mutated genes | Smoker | Non-smoker | |||
|---|---|---|---|---|---|
| Cases | Rate (%) | Cases | Rate (%) | ||
| EGFR | 22 | 44.90 | 27 | 55.10 | |
| ALK | 6 | 85.71 | 1 | 14.29 | |
| ROS1 | 1 | 25.00 | 3 | 75.00 | |
| MET | 3 | 75.00 | 1 | 25.00 | |
| ERBB2 (Her2) | 1 | 25.00 | 3 | 75.00 | |
| BRAF | 0 | 0 | 2 | 100.00 | |
| KRAS | 7 | 87.50 | 1 | 12.50 | |
Table 9
| Mutated genes | Smoker | Non-smoker | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Total | 37 | 32.74 | 34 | 30.09 | 0.0685 | |
| EGFR | 22 | 32.84 | 27 | 58.70 | 0.0064 | |
| ALK | 6 | 8.96 | 1 | 2.17 | 0.2375 | |
| ROS1 | 1 | 1.49 | 3 | 6.52 | 0.3023 | |
| MET | 3 | 4.48 | 1 | 2.17 | 0.6446 | |
| ERBB2 (Her2) | 1 | 1.49 | 3 | 6.52 | 0.3023 | |
| BRAF | 0 | 0 | 2 | 4.35 | 0.1636 | |
| KRAS | 7 | 10.45 | 1 | 2.17 | 0.1389 | |
Stage distribution of mutated NSCLC patients and stage differences in patients with mutated NSCLC
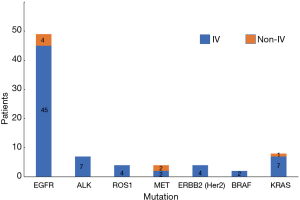
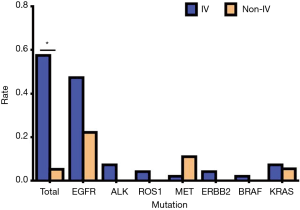
Of a total of 113 NSCLC cases, 78 people had genetic mutations, including 71 cases of stage IV and 7 cases of stage non-IV. In our study, the vast majorities of patients were advanced lung cancer cases, which might have been related to the source of the cases (Table 10, Figure 10). Our results showed that the total mutation rate was 57.52% in patients of stage IV and 5.31% in patients of the non-IV stage (P<0.05), demonstrating that the gene mutation rate in patients of stage IV was significantly higher than that in non-IV stage patients. In single gene mutations, there was no significant difference between stage IV and non-IV (P>0.05) (Table 11, Figure 11).
Table 10
| Mutated genes | IV | Non-IV | |||
|---|---|---|---|---|---|
| Cases | Rate (%) | Cases | Rate (%) | ||
| EGFR | 45 | 91.84 | 4 | 8.16 | |
| ALK | 7 | 100.00 | 0 | 0 | |
| ROS1 | 4 | 100.00 | 0 | 0 | |
| MET | 2 | 50.00 | 2 | 50.00 | |
| ERBB2 (Her2) | 4 | 100.00 | 0 | 0 | |
| BRAF | 2 | 100.00 | 0 | 0 | |
| KRAS | 7 | 87.50 | 1 | 12.50 | |
Table 11
| Mutated genes | IV | Non-IV | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Total | 65 | 57.52 | 6 | 5.31 | 0.0105 | |
| EGFR | 45 | 47.37 | 4 | 22.22 | 0.0864 | |
| ALK | 7 | 7.37 | 0 | 0 | 0.5949 | |
| ROS1 | 4 | 4.21 | 0 | 0 | 1.0000 | |
| MET | 2 | 2.11 | 2 | 11.11 | 0.1186 | |
| ERBB2 (Her2) | 4 | 4.21 | 0 | 0 | 1.0000 | |
| BRAF | 2 | 2.11 | 0 | 0 | 1.0000 | |
| KRAS | 7 | 7.37 | 1 | 5.56 | 1.0000 | |
Distribution of mutation types in NSCLC patients with EGFR gene mutation and association with clinical features
Of a total of 113 NSCLC cases, 78 cases had genetic mutations, with 49 of these being EGFR mutations. Among the 49 cases of EGFR mutation, there were 25 males (51.02%) and 24 females (48.98%). In addition, there were 22 people aged 60 or above (44.90%) and 27 people aged lower than 60 (55.10%). According to the 8th edition of TNM classification, there were 45 patients of stage IV (91.84%) and 4 patients of non-stage IV (8.16%). In 49 cases, 22 had a history of smoking. Histological distribution was characterized by 47 cases of ADC (95.92%) and 2 cases of SCC (4.08%). The distribution of each mutant type of EGFR mutation was as follows:
Exon 2, 1.35%; exon 4, 1.35%; exon 6, 1.35%; exon 18, 1.35%; exon 19, 33.78%; exon 20, 16.22%; exon 21, 25.68%; exon 22, 1.35%; and EGFR amplification, 17.57%.
Clearly, mutations in EGFR are mainly concentrated in exons 19, 20, and 21. However, EGFR amplification cannot be ignored (Table 12, Figure 12).
Table 12
| Clinicopathological features | Classification | Cases | Rate (%) |
|---|---|---|---|
| Gender | Male | 25 | 51.02 |
| Female | 24 | 48.98 | |
| Age | ≥60 | 22 | 44.90 |
| <60 | 27 | 55.10 | |
| TNM staging | IV | 45 | 91.84 |
| Non-IV | 4 | 8.16 | |
| Smoking history | Yes | 22 | 44.90 |
| No | 27 | 55.10 | |
| Pathological type | ADC | 47 | 95.92 |
| SCC | 2 | 4.08 | |
| EGFR gene mutations | Exon 2 | 1 | 1.35 |
| Exon 4 | 1 | 1.35 | |
| Exon 6 | 1 | 1.35 | |
| Exon 18 | 1 | 1.35 | |
| Exon 19 | 25 | 33.78 | |
| Exon 20 | 12 | 16.22 | |
| Exon 21 | 19 | 25.68 | |
| Exon 22 | 1 | 1.35 | |
| EGFR amplification | 13 | 17.57 |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Analysis of each EGFR mutation type
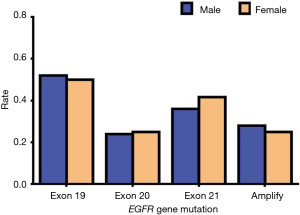
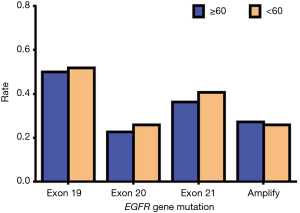
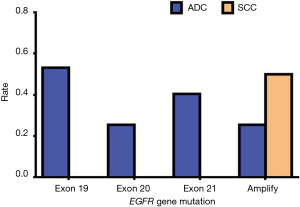
Since EGFR mutations are mainly concentrated in exons 19, 20, and 21 mutations and EGFR amplification, we mainly describe these types in detail. Of the exon 19 mutations of EGFR, males accounted for 13 of the total cases and females accounted for 12; there were 11 patients aged 60 or above and 14 patients aged lower than 60, and 25 patients were stage IV. Of the exon 19 mutations of EGFR, there were twelve patients who had a history of smoking, and all these cases were ADC. In exon 20 mutations of EGFR, males accounted for 6 of the total cases and females accounted for 6; there were 5 people aged 60 or above and 7 people aged lower than 60, and 12 patients of stage IV. In exon 20 mutations of EGFR, there were five patients who had a history of smoking, and all these cases were ADC. In exon 21 mutations of EGFR, males accounted for 9 of the total cases and females accounted for 10; there were 8 people aged 60 or above and 11 people aged lower than 60, and 16 patients of stage IV. In exon 21 mutations of EGFR, there were seven patients who had a history of smoking, and all these cases were ADC. In EGFR amplification mutations of EGFR, males accounted for 7 of the total cases, and females accounted for 6; there were 6 people aged 60 or above and 7 aged lower than 60, and 11 patients of stage IV. In EGFR amplification mutations of EGFR, there were eight patients who had a history of smoking, and only one case was SCC. According to the above data, we found that these types of mutations in EGFR had no apparent association with gender, age, and smoking history. Also, these types of mutations in EGFR are almost all advanced ADC (Table 13). Our results suggest that there is no statistically significant difference between exon 19, 20, and 21 mutations and EGFR gene amplification and clinical features (P>0.05) (Tables 14-18, Figures 13-17).
Table 13
| Clinicopathological features | Classification | EGFR mutation types | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 2 | Exon 4 | Exon 6 | Exon 18 | Exon 19 | Exon 20 | Exon 21 | Exon 22 | Amplification | ||
| Gender | Male | 1 | 1 | 0 | 1 | 13 | 6 | 9 | 1 | 7 |
| Female | 0 | 0 | 1 | 0 | 12 | 6 | 10 | 0 | 6 | |
| Age | ≥60 | 1 | 1 | 0 | 1 | 11 | 5 | 8 | 1 | 6 |
| <60 | 0 | 0 | 1 | 0 | 14 | 7 | 11 | 0 | 7 | |
| TNM staging | IV | 1 | 1 | 1 | 0 | 25 | 12 | 16 | 1 | 11 |
| Non-IV | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 2 | |
| Smoking history | Yes | 1 | 1 | 0 | 1 | 12 | 5 | 7 | 1 | 8 |
| No | 0 | 0 | 1 | 0 | 13 | 7 | 12 | 0 | 5 | |
| Pathological type | ADC | 1 | 1 | 1 | 0 | 25 | 12 | 19 | 1 | 12 |
| SCC | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Table 14
| Gene mutation types | Male | Female | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Exon 19 | 13 | 52.00 | 12 | 50.00 | 0.8887 | |
| Exon 20 | 6 | 24.00 | 6 | 25.00 | 0.8019 | |
| Exon 21 | 9 | 36.00 | 10 | 41.67 | 0.9095 | |
| Amplification | 7 | 28.00 | 6 | 25.00 | 0.9316 | |
Table 15
| Gene mutation types | ≥60 years | <60 years | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Exon 19 | 11 | 50.00 | 14 | 51.85 | 0.8974 | |
| Exon 20 | 5 | 22.73 | 7 | 25.93 | 0.9402 | |
| Exon 21 | 8 | 36.36 | 11 | 40.74 | 0.9856 | |
| Amplification | 6 | 27.27 | 7 | 25.93 | 0.8266 | |
Table 16
| Gene mutation types | IV | Non-IV | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Exon 19 | 25 | 55.56 | 0 | 0 | 0.0502 | |
| Exon 20 | 12 | 26.67 | 0 | 0 | 0.5599 | |
| Exon 21 | 16 | 35.56 | 3 | 75.00 | 0.2848 | |
| Amplification | 11 | 24.44 | 2 | 50.00 | 0.2839 | |
Table 17
| Gene mutation types | Smoker | Non- smoker | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Exon 19 | 12 | 54.55 | 13 | 48.15 | 0.8742 | |
| Exon 20 | 5 | 22.72 | 7 | 25.93 | 0.9402 | |
| Exon 21 | 7 | 31.82 | 12 | 44.44 | 0.5435 | |
| Amplification | 8 | 36.36 | 5 | 18.52 | 0.2792 | |
Table 18
| Gene mutation types | ADC | SCC | P value | |||
|---|---|---|---|---|---|---|
| Cases | Mutation rate (%) | Cases | Mutation rate (%) | |||
| Exon 19 | 25 | 53.19 | 0 | 0 | 0.2347 | |
| Exon 20 | 12 | 25.53 | 0 | 0 | 1.0000 | |
| Exon 21 | 19 | 40.43 | 0 | 0 | 0.5153 | |
| Amplification | 12 | 25.53 | 1 | 50.00 | 0.4643 | |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.


Discussion
Lung cancer is the most common malignancy in the world and is the leading cause of cancer death (23). Eighty-five percent of all lung cancer cases are NSCLC (24). Most patients with NSCLC were diagnosed as an advanced stage and lost the opportunity for radical surgery, yet targeted therapy based on gene mutations has significantly improved progression-free survival in patients with advanced NSCLC (25). In recent years, the improvement of gene detection efficiency and the application of tyrosine kinases inhibitors (TKIs) have greatly improved the therapeutic effect of lung cancer (26). Previous studies have suggested that most cases of NSCLC patients only express one type of gene mutation, each gene mutation population has different clinicopathological features, and each is expressed differently in different regions of the population (17). This study analyzed multiple gene mutations and clinical features of NSCLC patients in the Hunan region to learn its relevance. In our study, we found that of a total of 113 NSCLC cases, 78 people had genetic mutations. The distribution of each of the mutated genes was as follows: EGFR, 62.82%; ALK, 8.97%; ROS1, 5.13%; MET, 5.13%; ERBB2, 5.13%; RET, 0.00%; BRAF, 2.56%; KRAS, 10.26%. It is clear from the data that EGFR mutations are the most common.
Xiao et al. found that the rate of EGFR mutations in women was significantly higher than that in men and that the EGFR mutation rate in ADC was significantly higher than that in SCC (27). Paez et al. analyzed the mutation and clinical features of 119 patients with NSCLC, finding that EGFR mutations in ADC were higher than in other pathological types, with the chance of EGFR mutation is more frequent in women than in men (7). In addition, past studies have shown that in the factor analysis of smoking status, non-smokers EGFR mutation rate was 38.2%, which was far higher than the smokers mutation rate of 10.1%; EGFR mutation and the age of the patients with lung cancer and clinical stage had no obvious relation in these studies (28). As shown in our study, EGFR mutations were common in non-smoking women with ADC and had no obvious correlation with age. This is consistent with previous research showing that EGFR has the same mutation characteristics in different regions of the population. However, in our study, the mutation rate was higher in patients with stage IV. This differs from previous studies and may be related to the source of the cases.
ALK-mutated NSCLC accounts for 2% to 5% of all NSCLC cases (29). Compared with NSCLC patients with no gene mutation, ALK-positive NSCLC patients tend to be mild or never-smokers, young, male, and in a more advanced stage or stage IV disease. In histology, the NSCLC cases with ALK mutations are more frequent in ADC (30-32). As shown in our study, ALK mutations were more likely to occur in young men with stage IV of ADC. These results are similar to previous findings, but there is no significant difference, which may be related to the small sample size. ALK mutation cases tended to be smokers, which is different from previous studies and may be related to the sample size and regional population.
The mutation rate of ROS1 in NSCLC was 1.8% (33). ROS1 mutation usually occurred in ADC and was mainly observed in young, non-smoking, advanced stage, female patients (33-35). Our results were similar with previous studies of ROS1 mutations mainly observed in non-smoking, stage IV, female patients. However, in terms of age and organization type, our data demonstrated that more ROS1 mutations were detected in patients who were older or had SCC. There is no significant difference in the above data, which may be related to the small number of ROS1 mutation cases.
As for NSCLC, until 2007, there was only one report on the amplification of the MET gene (36). Recent studies have shown that NSCLC patients also report a similarly low incidence of MET gene amplification (1.4–11.1%) (37). However, a study from France showed that the incidence of the MET gene mutation was much higher (21%) (38). MET mutations are common in patients with advanced lung cancer. In another study, the MET mutations were found not to be significantly correlated with gender and age and were more prone to be SCC and smokers, although the differences were not significant (39). Similarly to this study, our results indicate that MET mutations are not significantly correlated with gender, age, pathological type, and smoking. This suggests that there may be no correlation between the MET mutations and these clinical features, or that the sample size of the MET mutation rate is low. The results of our study differ from previous studies in terms of the effects of staging on MET mutations, which may be related to our sample size.
Tomizawa et al. found that HER2 mutations are more common in women, non-smokers, and ADC (40). And Sonobe et al. found that ERBB2 mutations are more common in older, non-smoking, female, ADC, and early-stage cases (41). Our results showed that ERBB2 mutations were more common in women, ADC, and non-smokers, although there was no statistical difference. This is the same as the previous study. The lack of a significant difference can be attributed mainly to the low mutation rate of ERBB2 and the small number of cases. In our results, the more frequent ERBB2 mutations in young, late-stage lung cancer patients may be related to our small sample size or regional differences in population.
The frequency of BRAF mutations in NSCLC is about 4% (42,43). BRAF mutation is mainly found in ADC patients and was not significantly different in age, sex distribution, smoking history, and staging (44). Our results showed that BRAF mutation was not significantly correlated with gender, age, smoking history, and stage. This is the same as the previous study. The difference is that our study showed no significant difference in pathological types from BRAF mutations, but our two BRAF mutations are ADC cases and are considered to relate with a small number of cases. In short, it proves the clinical situation of BRAF mutation, and it proves the low mutation rate of BRAF.
Zhao et al. found that KRAS mutations were more common in men with ADC (45), while another study showed that KRAS mutations are more common in smokers (46). Our results showed that KRAS mutations are more common in men, smokers, older ages, SCC and advanced lung cancer cases, but none showed a statistical difference. Of these factors, male sex and smoking were consistent with other studies in their relation to KRAS mutation. In the type of pathology, age, and staging were different from other studies. It may be related to our study population comes from people who have already done genetic testing.
The most common EGFR mutations occur in exon 19 (about 46% of EGFR mutations), exon 20 (about 9% of EGFR mutations), and exon 21 (about 39% of EGFR mutations). The remaining 6% of the mutations occur mainly in exon 18 and other exons (47,48). EGFR amplification, as a kind of EGFR mutation, may be related to the progress of lung cancer (49). Soh et al. found that gene amplification may be related to the pathogenesis of advanced cancer (50). Zhang et al. found that the incidence of EGFR amplification in EGFR mutations was about 18%, and EGFR gene mutations occurred more frequently in samples that were accompanied by gene amplification than in those with high polysomy (51). Similar to previous studies, mutations in EGFR were mainly concentrated in exons 19, 20, and 21. Furthermore, EGFR amplification accounting for 17.57% is a type of mutation that cannot be ignored. It can serve as an important target for targeted therapy for NSCLC. Finally, there was no statistically significant difference between the mutations of exon 19, 20, 21, gene amplification and clinical features.
Conclusions
(I) EGFR mutation was more common in non-smoking female patients with ADC and had no significant correlation with age and stage. (II) EGFR mutations were mainly concentrated in exon 19, 20, and 21 and EGFR amplification. There was no significant statistical difference between mutations in exons 19, 20, 21, EGFR gene amplification and clinical features. (III) There was no statistically significant difference in the ALK/ROS1/MET/ERBB2/BRAF/KRAS mutations with gender, age, tissue type, smoking history, and tumor stage.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent from all patients and the approval from the Medical Ethics Committee of Xiangya Hospital of Central South University have been obtained (the ID number is 201512547).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Travis WD, Brambilla E, Riely GJ. New Pathologic Classification of Lung Cancer: Relevance for Clinical Practice and Clinical Trials. J Clin Oncol 2013;31:992-1001. [Crossref] [PubMed]
- Schiller JH, Harrington D, Belani CP, et al. Comparison of Four Chemotherapy Regimens for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Wang S, Peng L, Li J, et al. A trial-based cost-effectiveness analysis of erlotinib alone versus platinum-based doublet chemotherapy as first-line therapy for Eastern Asian nonsquamous non-small-cell lung cancer. PLoS One 2013;8:e55917. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Travis WD. Pathology of Lung Cancer. Clin Chest Med 2011;32:669-92. [Crossref] [PubMed]
- Yang JC, Sequist LV, Zhou C, et al. Effect of dose adjustment on the safety and efficacy of afatinib for EGFR mutation-positive lung adenocarcinoma: post hoc analyses of the randomized LUX-Lung 3 and 6 trials. Ann Oncol 2016;27:2103-10. [Crossref] [PubMed]
- Xia N, An J, Jiang QQ, et al. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res 2013;39:328-35. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Qiao X, Ai D, Liang H, et al. Gene Expression and Clinical Characteristics of Molecular Targeted Therapy in Non-small Cell Lung Cancer Patients in Shandong. Zhongguo Fei Ai Za Zhi 2017;20:14-20. [PubMed]
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [Crossref] [PubMed]
- Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. [Crossref] [PubMed]
- Dong QG, Han BH, Huang JS, et al. Analysis of EGFR mutations in 176 cases of non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi 2006;28:686-90. [PubMed]
- Zhang K, Chen H, Wang Y, et al. Clinical characteristics and molecular patterns of RET-rearranged lung cancer in Chinese patients. Oncol Res 2019;27:575-82. [Crossref] [PubMed]
- Feng Y, Feng G, Lu X, et al. Exploratory analysis of introducing next-generation sequencing-based method to treatment-naive lung cancer patients. J Thorac Dis 2018;10:5904-12. [Crossref] [PubMed]
- Soerjomataram I, Lortet-Tieulent J, Parkin DM, et al. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 2012;380:1840-50. [Crossref] [PubMed]
- Felip E, Stahel RA, Pavlidis N. ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of non-small-cell lung cancer (NSCLC). Ann Oncol 2005;16:i28-9. [Crossref] [PubMed]
- Lu RL, Hu CP, Yang HP, et al. Biological characteristics and epidermal growth factor receptor tyrosine kinase inhibitors efficacy of EGFR mutation and its subtypes in lung adenocarcinoma. Pathol Oncol Res 2014;20:445-51. [Crossref] [PubMed]
- Wang S, Wang Z. EGFR mutations in patients with non-small cell lung cancer from mainland China and their relationships with clinicopathological features: a meta-analysis. Int J Clin Exp Med 2014;7:1967-78. [PubMed]
- Xiao D, Lu C, Zhu W, et al. Comparison of small biopsy specimens and surgical specimens for the detection of EGFR mutations and EML4-ALK in non-small-cell lung cancer. Oncotarget 2016;7:59049-57. [Crossref] [PubMed]
- Liu H, Li Y, Chen G, et al. Detection and Its Clinical Significance of EGFR Gene Mutation and Gene Amplification in 187 Patients with Non-small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi 2009;12:1219-28. [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216-23. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Go H, Kim DW, Kim D, et al. Clinicopathologic Analysis of ROS1-Rearranged Non-Small-Cell Lung Cancer and Proposal of a Diagnostic Algorithm. J Thorac Oncol 2013;8:1445-50. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nature Medicine 2012;18:378. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ignatius Ou SH, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res 2005;65:5561-70. [Crossref] [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [Crossref] [PubMed]
- Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol 2008;3:331-9. [Crossref] [PubMed]
- Chen YT, Chang JW, Liu HP, et al. Clinical implications of high MET gene dosage in non-small cell lung cancer patients without previous tyrosine kinase inhibitor treatment. J Thorac Oncol 2011;6:2027-35. [Crossref] [PubMed]
- Tomizawa K, Suda K, Onozato R, et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer 2011;74:139-44. [Crossref] [PubMed]
- Sonobe M, Manabe T, Wada H, et al. Lung Adenocarcinoma Harboring Mutations in the ERBB2 Kinase Domain. J Mol Diagn 2006;8:351-6. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [Crossref] [PubMed]
- Cardarella S, Ogino A, Nishino M, et al. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res 2013;19:4532-40. [Crossref] [PubMed]
- Zhao J, Gao J, Guo L, et al. EGFR and KRAS Gene Mutations in 754 Patients with Resectable Stage I-IIIa Non-small Cell Lung Cancer and Its Clinical Significance. Zhongguo Fei Ai Za Zhi 2017;20:617-22. [PubMed]
- Nelson HH, Christiani DC, Mark EJ, et al. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. J Natl Cancer Inst 1999;91:2032-8. [Crossref] [PubMed]
- Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 2007;25:587-95. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Yatabe Y, Takahashi T, Mitsudomi T. Epidermal growth factor receptor gene amplification is acquired in association with tumor progression of EGFR-mutated lung cancer. Cancer Res 2008;68:2106-11. [Crossref] [PubMed]
- Soh J, Toyooka S, Ichihara S, et al. Sequential molecular changes during multistage pathogenesis of small peripheral adenocarcinomas of the lung. J Thorac Oncol 2008;3:340-7. [Crossref] [PubMed]
- Zhang LJ, Cai L, Li Z, et al. Relationship between epidermal growth factor receptor gene mutation and copy number in Chinese patients with non-small cell lung cancer. Chin J Cancer 2012;31:491-9. [Crossref] [PubMed]