
The hypermethylation of the CDKN2A and CHFR promoter region is a key regulatory mechanism of CDKN2A and CHFR expression in esophageal squamous cell carcinoma
Introduction
Esophageal cancer ranks sixth in mortality among all cancers worldwide (1,2). Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of esophageal cancer in East Asian countries (3). ESCC is generally treated using multimodal approaches, including surgery, radiotherapy and chemotherapy, and the overall 5-year survival rate ranges from 15% to 25% (2). The outcomes in patients with ESCC are related to factors including stage of diagnosis, the propensity for metastasis, radiosensitization and chemosensitivity. Radiotherapy is the standard treatment for patients with inoperable or locally advanced ESCC (4), and has played an important role in the management of esophagectomies as a neoadjuvant or adjuvant therapy. Although radiotherapy can be a useful palliative treatment for the clinical symptoms of ESCC, sustained remission and long-term survival are rarely achieved. In locally advanced ESCC, radiotherapy without chemotherapy can achieve a long-term survival rate of around 2–20% overall survival (OS) after 5 years (5). Radiosensitization and radioresistance are key factors affecting therapeutic efficacy.
With increasing radiosensitization, modulating the DNA damage response has been a long-standing issue in translational radiotherapy research. Several cell cycle checkpoints allow the repair of, and prevent the propagation of, damaged DNA (6). The failure of cell cycle checkpoint functions causes predisposed cells to undergo neoplastic transformation. Many cancers have an impaired DNA damage response and defective cell cycle checkpoints, as exemplified by the high incidence of ataxia-telangiectasia mutated (ATM), tumor protein p53 (TP53) and cyclin-dependent kinase inhibitor 2A (CDKN2A) mutations (7).
The CDKN2A gene is located within the chromosomal region 9 of p21 (8). CDKN2A encodes an unrelated tumor suppressor protein, an alternate open reading frame which interacts with the p53 regulatory protein, mouse double minute 2 homolog (MDM2) (9). The genomic DNA methylation of CDKN2A has been found to cause inactivation in various cancers, including lymphoma (2), head and neck cancers (10), gastric cancer (11), lung cancer (12) and esophageal cancer. In ESCC, the promoter region of CDKN2A has been found to be highly methylated in tissue samples from Chinese subjects (81.7%; 210/257) (13). Additionally, the expression of CDKN2A may increase the radiosensitivity of head and neck squamous cell carcinoma (HNSCC) cells (14).
The checkpoint protein with FHA and RING finger domains (CHFR) gene is a RING-type E3 ubiquitin (Ub) ligase, which regulates numerous important cellular proteins, and thus functions as a mitotic checkpoint and tumor suppressant (15-17). CHFR elevates p53 acetylation by destabilizing Silent mating type information regulation 2 homolog- 1 (SIRT1), resulting in an increase in its transcriptional activity and apoptotic cell death (18). In cancer, CHFR is more frequently inactivated than all other mitotic checkpoint control genes (19). Previous studies have shown that approximately 40% of tumors have reduced CHFR expression (95% CI: 26–54%) (20). In colorectal cancer and gastric cancer, CHFR inactivation has been found to be associated with microsatellite instability (MSI) and MLH1 promoter CpG island methylation (21). CpG island hypermethylation of the promoter region in the CHFR gene is also a key mechanism involved in silencing the CHFR gene in patients with esophageal cancer (22,23).
More than 50% of cancer patients receive radiotherapy at some time during the course of their disease. However, tumor cells have been shown to acquire radioresistance, which has been linked to increased rates of recurrence and treatment failure. The exact mechanisms by which tumor cells develop an adaptive resistance to therapeutic fractional irradiation remain unknown (24).
The objective of the present study was to evaluate the correlation between the expression of CDKN2A and CHFR, and the methylation status of the CpG island in the promoter region of (I) the ESCC cell line; (II) the radioresistant ESCC cell line; and (III) ESCC clinical tissue specimens. The role of CDKN2A and CHFR gene expressions in the radioresistance of ESCC was also examined. The study was approved by the Ethics Committee of The First Affiliated Hospital of USTC (Anhui Provincial Hospital) (2018-KY-22) and conforms to the provisions of in accordance with the Helsinki Declaration as revised in 2013.
Methods
Cell culture and reagents
The KYSE-150 and KYSE-180 ESCC cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. The radioresistant ESCC cell lines were irradiated by X-ray radiation, with a dose of 8 Gy, using a Varian CX linear accelerator. The distance between the radiation source and the specimens was 100 cm, the area of the radiation field was 20 cm × 20 cm and the X-ray absorbed dose rate was 1.50 Gy/min. Following X-ray irradiation, the ESCC cell lines were cultured in RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum, in an atmosphere containing 5% CO2 at a temperature of 37 °C, until the radioresistant cell clone was established and proved to be stable. The radioresistant ESCC cell lines derived from the KYSE-150 and KYSE-180 ESCC cell lines were denoted KYSE-150-radioresistance and KYSE-180-radioresistance, respectively.
Patients and samples
Twenty-eight samples of esophageal tissue were collected from patients diagnosed with histologically confirmed ESCC. All patients were recruited from the First Affiliated Hospital of the University of Science and Technology of China (USTC), between May 2014 and April 2015. All tissue samples were diagnosed histologically as ESCC, before being frozen immediately after resection and stored in liquid nitrogen. None of the patients underwent preoperative treatment (e.g., chemotherapy or radiation treatment). Specimens were classified histologically using the seventh edition of the UICC staging system for esophageal cancer. All subjects gave their informed consent for inclusion before participating in the study. The clinical characteristics of the patients are shown in Table 1.
Table 1
| Characteristics | Values |
|---|---|
| Sex (male/female) | 25/3 |
| Age (year), mean ± SD [range] | 63.32±9.38 [39–80] |
| TNM stage* | |
| I–II | 23 |
| III | 5 |
*, according to the UICC/AJCC TNM classification (seventh edition).
DNA isolation, bisulfate conversion and pyrosequencing
DNA was extracted from esophageal tissue and cell lines by proteinase K digestion and using a tissue DNA extraction kit (Qiagen Co., Hilden, Germany), according to the manufacturer’s instructions. Bisulfite modification of DNA (1 µg) was performed using an EpiTect Plus DNA Bisulfite Kit (Qiagen Co., Hilden, Germany), according to the manufacturer’s instructions. All purified genomic DNA samples were successfully tested by polymerase chain reaction with human β-actin primers. Bisulfite-modified gene promoters were amplified from genomic DNA (0.4 µg) via PCR. The PCR conditions were as follows: a temperature of 95 °C for 15 min; a temperature of 94 °C for 30 s; an annealing temperature (56 °C) for 30 s; a temperature of 72 °C for 30 s, followed by 45 cycles; and ending with an extension at a temperature of 72 °C for 10 min. The PCR product was sequenced using a pyrophosphate sequencing machine and PyroMark Gold Q96SQA Reagents (Qiagen Co., Hilden, Germany), according to the manufacturer’s instructions.
Quantitative real-time PCR
mRNA was reverse-transcribed into complementary DNA (cDNA) in a 200 µL reaction volume containing the following reagents: 15 µg of total mRNA; 5 µmol/L oligodT; 0.5 mmol/L dNTPs; 40 µL of 5×buffer; 10 mmol/L DL-Dithiothreitol (DTT); 400 units of Ribonuclease (RNase) inhibitor; 2,000 units of Moloney Murine Leukemia Virus (M-MLV); and distilled water (ultrapure; DNase and RNase free). The primers for each gene detected by real-time PCR are given in Table 2. The reverse transcription (RT) reaction was performed at a temperature of 37 °C for 50 minutes, and subsequently heated at a temperature of 70 °C for 15 minutes. The standard 50 µL volume reaction contained 25 µL 2× PCR buffer, 2 µL cDNA template and 0.4 µmol/L forward and reverse primers. Quantitative real-time PCR was performed using a Roche Light Cycler 480 II instrument (Roche Diagnostics, Germany). PCR reactions were performed using a total of 45 cycles, each consisting of a 15-s melt step at a temperature of 95 °C, followed by a 30-s annealing step at a temperature of 60 °C and a 30-s extension step at a temperature of 72 °C. Each sample was analyzed in triplicate for each target gene.
Table 2
| Genes | Size | Forward primer | Reverse primer |
|---|---|---|---|
| CHFR | 295 | ataccagcaccagtggaaca | tcctcaaagcacacctgagt |
| CDKN2A | 357 | aacgcaccgaatagttacgg | tcaatcggggatgtctgagg |
| β-actin | 101 | TTGCCGACAGGATGCAGAA | GCCGATCCACACGGAGTACTT |
Immunohistochemical assay of CDKN2A and CHFR protein expression
Twenty-eight ESCC samples was immunohistochemically stained by the Envision method. The sections (5 µm thick) were deparaffinized, rehydrated with graded concentrations of ethanol, and incubated with 3% H2O2 for 15 min. The sections were then incubated with the primary antibody (rabbit anti-human CHFR monoclonal antibodies, BS-4272R, Bioss, Beijing, China; mouse anti-human p16 monoclonal antibodies, ZM-0205, Zsbio, Beijing, China), followed by incubation with secondary antibodies (anti-mouse/rabbit, PV-6000, Zsbio, Beijing, China) for 15 min at room temperature and DAB reagent (ZLI-9018, Zsbio, Beijing, China).The sections were then counterstained with hematoxylin, dehydrated. CHFR, and CDKN2A (p16) staining were scored for nuclear and cytoplasmic staining based on intensity and percentage of cells stained (25,26).
Estimates of overall survival for ESCC patients for different levels of CDKN2A and CHFR genes expression
Follow-up analyses were conducted for all patients after esophagectomy, at intervals ranging from 8 to 42 months (median: 28.9 months), in order to evaluate the overall survival for ESCC patients for different levels of CDKN2A and CHFR expression. Overall survival outcome was classified as either survival or death.
Statistical analysis
Pyrosequencing data were analyzed using PyroMark CpG Assays (Qiagen Co., Hilden, Germany). A Mann-Whitney U test was performed to compare the methylation levels of CDKN2A and CHFR of every CpG site, in both the ESCC group and the control group. The association between the CpG site methylation and the clinicopathologic parameters was evaluated by the K independent samples test (media test). Spearman’s correlation was used to evaluate the correlations between the CpG site methylation levels of CDKN2A and CHFR and their expression levels. All P values were two sided, and their significance level was P<0.05.
Results
In radioresistant ESCC cell lines, the methylation of CDKN2A and CHFR genes was enhanced, and the expression of CDKN2A and CHFR genes was correspondingly decreased
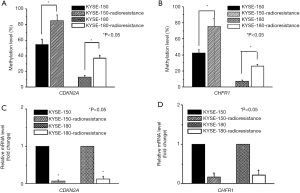
We examined the methylation and expression of CDKN2A and CHFR in ESCC cell lines and radioresistant ESCC cell lines. The measured methylation levels of CDKN2A were 54.59% and 12.68% in KYSE-150 and KYSE-180, respectively, and 85.22% and 36.96% in KYSE-150-radioresistance and KYSE-180-radioresistance, respectively (P<0.05) (Figure 1A). The measured methylation levels of CHFR were 42.33% and 7.36% in KYSE-150 and KYSE-180, respectively, and 75.22% and 25.93% in KYSE-150-radioresistance and KYSE-180-radioresistance, respectively (P<0.05) (Figure 1B). When we compared the levels of CDKN2A mRNA and CHFR mRNA in the ESCC cell lines before and after irradiation, the expression of CDKN2A and CHFR was found to be significantly lower in the radioresistant ESCC cells lines than in the original cell lines (P<0.05) (Figure 1C,D).
The methylation of CDKN2A and CHFR genes was higher in the ESCC and paracarcinoma tissues than in the adjacent benign tissue, and the expression of CDKN2A and CHFR genes in both the ESCC and paracarcinoma tissues was correspondingly decreased
The measured methylation levels of CHFR were 46.32% in the ESCC tissue, 42.05% in the paracarcinoma tissue and 22.33% in the adjacent normal tissue. The measured methylation levels of CDKN2A were 27.51% in the ESCC tissue, 24.54% in the paracarcinoma tissue and 10.89% in the adjacent normal tissue. The measured methylation frequencies of CDKN2A and CHFR in the malignant and paracarcinoma tissues were significantly higher than in the adjacent benign tissue (Figure 2A). The measured CDKN2A and CHFR methylation was not correlated with the Union for International Cancer Control (UICC) stage in the malignant tissue, paracarcinoma tissue or in the adjacent benign tissue. In resected samples, the levels of CDKN2A mRNA and CHFR mRNA were lower in the ESCC and paracarcinoma tissues than they were in the adjacent normal tissue (Figure 2B).

The relationship between the expression of CDKN2A and CHFR genes in the ESCC tissue, paracarcinoma tissue and adjacent normal tissue in tumor staging and the degree of tumor differentiation
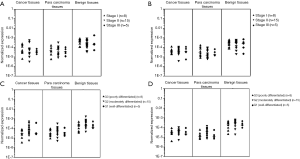
The levels of CDKN2A mRNA and CHFR mRNA in the ESCC tissue, paracarcinoma tissue and adjacent normal tissue were not correlated with UICC stage (Figure 3A,B). The mRNA transcription of CDKN2A was related to the histologic grade in G2 (moderately differentiated) and G3 (poorly differentiated) group of normal esophageal tissue P=0.026) (Figure 3C). The mRNA transcription of CHFR was related to the histologic grade in G2 (moderately differentiated) and G3 (poorly differentiated) group of adjacent cancerous tissue (P=0.034) (Figure 3D).
Correlation between CDKN2A and CHFR genes immunohistochemical expression and DNA methylation
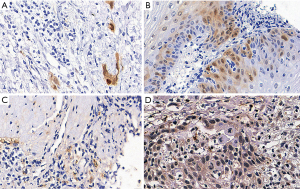
The average percentage of CDKN2A protein expression in hyper- and hypo-methylation groups immune-histochemically were 3.65% and 17.66% (Figure 4A,B). The average percentage of CHFR protein expression in hyper- and hypo-methylation groups immune-histochemically were 20.84% and 37.62% (Figure 4C,D).
Overall survival rates were significantly correlated with levels of CDKN2A and CHFR genes expression in ESCC patients
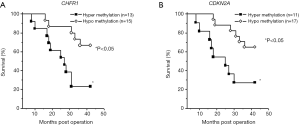
In the current study, we analyzed clinical follow-up data according to methylation levels. The median length of the follow-up period was 32 months. The methylation levels of CDKN2A and CHFR were not correlated with TNM staging (CDKN2A, P=0.298; CHFR, P=0.880). The overall survival rates were significantly correlated with the levels of CDKN2A and CHFR expression (hypermethylation: methylation level of CDKN2A >20%; methylation level of CHFR1 >40%; hypomethylation: methylation level of CDKN2A <20%; methylation level of CHFR1 <40%; P<0.05) (Figure 5).
Discussion
Despite numerous intensive recent studies aimed at improving the treatment of esophageal cancer, clinical outcomes remain unsatisfactory (2,8,9). Genetic and epigenetic changes have been found to accumulate during the development of various cancers, including ESCC. Aberrant DNA methylation affects genes involved in the cell cycle, DNA damage repair, and in Wnt, transforming growth factor-β (TGF-β) and nuclear factor-kappa B (NF-κB) signaling pathways (10). The identification of genes with aberrant methylation in accordance with the multi-stage process of ESCC could allow more accurate diagnosis, better prediction of the risk of radioresistance and better disease prognosis.
The CDKN2A tumor suppressor gene, which encodes the p16 INK4a protein, has been found to have a causal link with several different types of cancers, including lung cancer, head and neck cancer, gastric cancer, colorectal cancer and esophageal cancer. CHFR is a checkpoint gene. Failure of CHFR function results in genomic instability, a mutagenic condition that predisposes cells to undergo neoplastic transformation (11,12). A number of studies have identified the hypermethylation of the CpG island in the promoter region as a key mechanism involved in gene regulation (13,27,28).
In the present study, the expression levels of CDKN2A mRNA and CHFR mRNA were found to be lower in radioresistant ESCC cell lines than in normal ESCC cell lines, and decreases in CDKN2A and CHFR expression were found to be correlated with methylation of the CDKN2A and CHFR promoter region. This study indicates that promoter hypermethylation could be a key regulatory mechanism of CDKN2A and CHFR transcription, and play an important role in the radioresistance of ESCC.
This study found that the methylation frequencies of CDKN2A and CHFR in the malignant tissue and paracarcinoma tissue were significantly higher than those in the adjacent benign tissue. The levels of CDKN2A mRNA and CHFR mRNA were found to be frequently decreased in the ESCC tissue and the CDKN2A mRNA and CHFR mRNA levels in the ESCC tissue, paracarcinoma tissue and adjacent normal tissue were not found to be correlated with UICC stage. The average percentage of CDKN2A and CHFR protein expression in hyper-methylation groups were lower than that in hypo-methylation groups. The results indicate that the methylation of CDKN2A and CHFR could play an important role in the pathogenesis of ESCC. The methylation of the CHFR and CDKN2A genes could be related to a differentiated degree of ESCC, and could be an early molecular phenomenon in the disease.
Conclusions
The results of the present study indicate that the expression of CDKN2A and CHFR is frequently suppressed in ESCC, and that the hypermethylation of the CDKN2A and CHFR promoter region is a key regulatory mechanism involved in the expression of CDKN2A and CHFR in ESCC. Promoter hypermethylation and the downregulation of CDKN2A and CHFR in ESCC tissue may represent a promising biomarker for predicting the radioresistance of ESCC. However, the study was limited by a lack of direct functional analysis of CDKN2A and CHFR, and by the fact that it did not study the therapeutic potential of demethylation treatment in radioresistant ESCC cells. Further studies are therefore necessary to identify the direct function of CDKN2A and CHFR, and the ability of DNA demethylation to treat the radioresistance of ESCC.
Acknowledgments
Funding: The study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of USTC (Anhui Provincial Hospital) (2018-KY-22) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003;349:2241-52. [Crossref] [PubMed]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A 1993;90:11995-9. [Crossref] [PubMed]
- Chen H, Zhou L, Yang Y, et al. Clinical Effect of Radiotherapy Combined with Chemotherapy for Non-Surgical Treatment of the Esophageal Squamous Cell Carcinoma. Med Sci Monit 2018;24:4183-91. [Crossref] [PubMed]
- Lee E, Lee BB, Ko E, et al. Cohypermethylation of p14 in combination with CADM1 or DCC as a recurrence-related prognostic indicator in stage I esophageal squamous cell carcinoma. Cancer 2013;119:1752-60. [Crossref] [PubMed]
- Bertholon J, Wang Q, Falette N, et al. Chfr inactivation is not associated to chromosomal instability in colon cancers. Oncogene 2003;22:8956-60. [Crossref] [PubMed]
- Ando N, Ozawa S, Kitagawa Y, et al. Improvement in the results of surgical treatment of advanced squamous esophageal carcinoma during 15 consecutive years. Ann Surg 2000;232:225-32. [Crossref] [PubMed]
- Law S, Kwong DL, Kwok KF, et al. Improvement in treatment results and long-term survival of patients with esophageal cancer: impact of chemoradiation and change in treatment strategy. Ann Surg 2003;238:339-47; discussion 347-8. [PubMed]
- Ma K, Cao B, Guo M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin Epigenetics 2016;8:43. [Crossref] [PubMed]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science 1989;246:629-34. [Crossref] [PubMed]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science 1994;266:1821-8. [Crossref] [PubMed]
- Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 2001;61:3410-8. [PubMed]
- Dok R, Abbasi Asbagh L, Van Limbergen EJ, et al. Nuclear p16INK4a expression predicts enhanced radiation response in head and neck cancers. Oncotarget 2016;7:38785-95. [Crossref] [PubMed]
- Soutto M, Peng D, Razvi M, et al. Epigenetic and genetic silencing of CHFR in esophageal adenocarcinomas. Cancer 2010;116:4033-42. [Crossref] [PubMed]
- Kuwabara T, Hiyama T, Tanaka S, et al. Genetic pathways of multiple esophageal squamous cell carcinomas. Oncol Rep 2011;25:453-9. [PubMed]
- Qureshi MA, Jan N, Dar NA, et al. A novel p16(INK4A) mutation associated with esophageal squamous cell carcinoma in a high risk population. Biomarkers 2012;17:552-6. [Crossref] [PubMed]
- Yu X, Minter-Dykhouse K, Malureanu L, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet 2005;37:401-6. [Crossref] [PubMed]
- Kim JM, Cho EN, Kwon YE, et al. CHFR functions as a ubiquitin ligase for HLTF to regulate its stability and functions. Biochem Biophys Res Commun 2010;395:515-20. [Crossref] [PubMed]
- Oh YM, Kwon YE, Kim JM, et al. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol 2009;11:295-302. [Crossref] [PubMed]
- Toyota M, Sasaki Y, Satoh A, et al. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci U S A 2003;100:7818-23. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 2007;21:525-30. [Crossref] [PubMed]
- Ahmed KM, Li JJ. NF-kappa B-mediated adaptive resistance to ionizing radiation. Free Radic Biol Med 2008;44:1-13. [Crossref] [PubMed]
- Zhang D, Xu XL, Li F, et al. Upregulation of the checkpoint protein CHFR is associated with tumor suppression in pancreatic cancers. Oncol Lett 2017;14:8042-50. [PubMed]
- Satgunaseelan L, Virk SA, Lum T, et al. p16 expression independent of human papillomavirus is associated with lower stage and longer disease-free survival in oral cavity squamous cell carcinoma. Pathology 2016;48:441-8. [Crossref] [PubMed]
- Nie Y, Yang G, Song Y, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis 2001;22:1615-23. [Crossref] [PubMed]
- Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol 2000;183:145-54. [Crossref] [PubMed]


