
Overexpression and mutation of ZNF384 is associated with favorable prognosis in breast cancer patients
Introduction
The latest statistics suggest that breast cancer has become one of the greatest threats to women’s health worldwide, and its age of onset shows a younger trend (1,2). In China, the breast cancer burden is increasing rapidly accompanied by the elevation of its global proportion. Different from the moderate morbidity and decreased mortality in developed countries, the morbidity and mortality of breast cancer in China present an obvious increasing trend during the past several decades (3,4). Moreover, the survival rate of breast cancer in China is also lower than that in developed countries. In a word, the situation of breast cancer diagnosis, treatment and prevention is extremely serious (5,6). At the moment, research on the breast cancer prognosis-related markers remains in the initial stage. Some studies have reported that molecular markers such as miRNA-21 (7), miRNA-210 (8), CXCR4 (9), ALDH1A1 (10) and BRCA1/BRCA2 (11), are correlated with the prognosis for breast cancer, nonetheless, the precise underlying mechanism remains unclear. Thus, genetic markers should be further discovered and verified in much more studies, so as to explore their application value related to clinical prognosis, consequently providing important foundation and thinking for the preventive intervention, early diagnosis, treatment and prognostic evaluation of breast cancer.
This study had first reported the correlation of the survival of breast cancer patients with ZNF384, the fusion gene that was extensively reported to be involved in the genesis, development and malignant transformation of multiple leukemia subtypes (12,13), through the combined application of whole exome sequencing (WES) of clinical samples and multiple bioinformatics analysis approaches. Besides, the potential downstream molecular mechanism was also explored. Our results preliminarily verified that ZNF384 could serve as the molecular marker for diagnosis and prognosis judgement of breast cancer, which might potentially be a promising therapeutic target.
Methods
Source of clinical patients, WES and analysis
Tissue samples were collected from ten breast cancer patients treated by surgical operation in Department of Breast Surgery, Women’s Hospital of Zhejiang University School of Medicine with the approval of the Ethics Committee. The collected samples were sent to Beijing Novogene Science and Technology Co., Ltd for WES and subsequent analyses. The quality requirements of the sequencing data were as follows, average Q30 of >80% and error rate of <0.1%. Requirements for data analysis: (I) variation sites with the depth of <10× should be filtered; (II) SNP sites in the dbSNP database should be eliminated, but the variations in COSMIC database should be preserved; (III) sites in intergenic region, non-coding region, intron region and synonymous mutation should be removed; and (IV) variation sites in genomic repeat region should be ruled out.
Gene enrichment analysis
KEGG signaling pathway and disease level enrichment analysis: DAVID bioinformatics tools were adopted for enrichment analysis for the common mutant genes discovered from the multiple samples in WES (14). Meanwhile, the target genes were input into the database for further gene set analysis. At the same time, corresponding gene identifiers were selected, the human genome-wide was ticked as the background, “Functional annotation tool” was selected as the analytical tool, and the KEGG signaling pathway and disease level analysis was selected from the results.
GSEA enrichment analysis (15): the GSEA3.0 software was adopted for analysis. The c2.Cp.Kegg.V6.1.symbols.gmt dataset from MsigDB database was downloaded from GSEA website. Subsequently, the expression profile data stratified according to high and low expression, as well as the property files were conducted enrichment analysis according to the default weighted enrichment statistical method, and the force fitting frequency was set as 1,000.
Excavation of co-expression genes
The Multi-Experiment Matrix (MEM) software (16) was selected, the gene names were input, and the Affymetrix GeneChip Human Genome U133 Plus 2.0 dataset was selected for subsequent analysis.
The Cancer Genome Atlas (TCGA) data analysis
Breast cancer clinical data (including gene expression in cancer and para-carcinoma tissues, mutation level data and patient survival data) were downloaded from the TCGA database (17). The consistency analysis between gene expression level and mutation was performed using the cBioPortal software. Measurement data were analyzed using the SPSS 17.0 statistical software, while the Graphpad prism 6 software was adopted for plotting.
Data statistics
Data analysis was performed using SPSS 17.0 statistical software, and the Graphpad 6.0 software was used for plotting. Measurement data were expressed as mean ± SD, comparisons between two samples were analyzed using t test, one-way analysis of variance (ANOVA) was employed for comparisons among multiple samples, and Log-rank (Mantel-Cox) test was adopted for survival analysis. Difference with P<0.05 was deemed as statistically significant.
Results
Screening of common mutant genes in tissue samples from clinical breast cancer patients
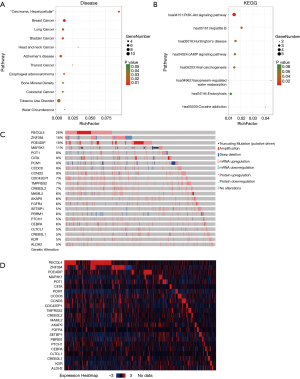
Tissue samples were collected immediately after resection from ten breast cancer patients admitted and treated in the Oncology Department in our hospital for WES, quality control and analysis. Our results suggested that, there were 23 common mutant genes among these samples. The common mutant genes existing in all the 10 samples were CCDC6, CCND3, CREB3L1, CREB3L2, MAML2, POT1, RECQL4 and CLTCL1. The common mutant genes in 9 samples were PBRM1, PCM1, ZNF384 and SETBP1. The common mutant genes in 8 samples were MAP3K1, AKAP9 and FGFR4. The common mutant genes in 7 samples were KDR and PTCH. The common mutant genes in 6 samples were CIITA and PDE4DIP. The common mutant genes in 5 samples were ALDH2, CDC42EP1, CEBPA and TMPRSS2. Afterwards, we applied the DAVID bioinformatics tools for functional annotations of these 23 genes. The results (Figure 1A,B) suggested that, most of these genes were enriched in multiple disease and signaling pathways such as breast cancer, liver cancer, lung cancer, head and neck cancer, endocytosis, hepatitis, PI3K/Akt and cAMP pathways.
ZNF384 was a potential predictive marker for the prognosis of breast cancer patients
To further confirm the changes in molecular pathological level and patient survival resulted from the mutations of the above 23 genes, the cBioPortal software was first selected for the consistency analysis between gene expression level and mutation of data extracted from the TCGA database (18). The results (Figure 1C,D) indicated that, among the 23 genes, only 2 genes showed high correlations and consistency between the expression changes and mutation (over 15%), which were ZNF384 and PDE4DIP. Afterwards, the breast cancer samples in TCGA database were analyzed. The results indicated that only ZNF384 mutation was correlated with the overall survival (OS, P=0.0352) and disease-free survival (DFS, P=0.0489) of breast cancer patients (Figure 2A,B,C,D). These findings revealed that, ZNF384 gene mutation in breast cancer tissues and cells might change its gene expression level to delay the course of disease and extend patient survival.
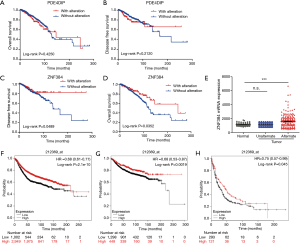
Then, the expression levels of ZNF384 gene among breast cancer tissues and paracancerous tissues with and without ZNF384 gene mutation were further compared. The results (Figure 2E) demonstrated that the expression level of ZNF384 gene in breast cancer tissues with ZNF384 mutation was far higher than that with no ZNF384 mutation (P<0.005), while no difference was observed between breast cancer tissues without ZNF384 gene mutation and normal tissues (P>0.005). These findings suggested that high ZNF384 expression was markedly correlated with mutation. Next, we analyzed the published microarray datasets through the Kaplan-Meier Plotter database (212369_at) (19). The results (Figure 2F,G,H) discovered that, high ZNF384 expression could outstandingly prolong the OS (HR =0.68, P=2.1×10−10), distal metastasis-free survival (HR =0.68, P=0.0019), and post-progression survival (HR =0.75, P=0.045). The above results proved that, ZNF384 mutation could up-regulate its expression level and extend the survival of breast cancer patients, which was thereby a potential predictive marker for the prognosis of breast cancer patients.
ZNF384 might regulate the CXCL14-mediated cell cycle and mitosis related pathways to exert the downstream effect
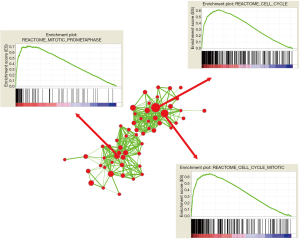
To further dig out the mechanism by which ZNF384 regulated the breast cancer process and affected patient survival, we carried out GSEA enrichment analysis of ZNF384 gene. The results (Figure 3) revealed that, samples with high ZNF384 expression were mainly enriched in the gene sets such as cell cycle (P=0.0031, FDR =0.075, enrichment score =−0.312), mitosis (P=0.021, FDR =0.131, enrichment score =−0.362) and mitochondrial pathway (P=0.0019, FDR =0.062, enrichment score =−0.431).
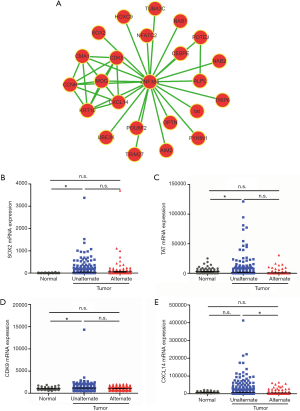
On this foundation, we then searched for the co-expression gene clusters with ZNF384 using the MEM software in order to further explore the downstream target gene of ZNF384. Moreover, we found out genes whose expression levels in breast cancer tissues were correlated with ZNF384 mutation from the gene clusters through analyzing the TCGA database (Figure 4A). The results (Figure 4B,C,D,E) indicated that only the expression level of CXCL14 was markedly changed in breast cancer tissues with ZNF384 mutation (down-regulated, P<0.05). All these results indicated that ZNF384 might suppress the CXCL14-mediated cell cycle and mitosis related pathways to delay breast cancer progression and promote patient survival.
Discussion
Breast cancer is one of the most common malignancies, with both incidence and mortality ranking first in all female malignant tumors. In China, breast cancer has severely threatened female health, with the annual new cases account for 12.2% of the global new cases, and the number of deaths took up 9.6% (20). Therefore, it is of great significance to discover the novel genesis and development-related molecular markers for us to enhance the diagnosis and treatment of breast cancer patients.
ZNF384 gene encodes a transcription factor, which had also been reported to regulate the levels of several extracellular matrix genes (including MMP1, MMP3, MMP7 and COL1A1) in multiple tumor types (21). Moreover, as a fusion gene, ZNF384 can fuse and rearrange with genes like EP300, CREBBP, ARID1B, SYNRG, EWSR1 and SMARCA2, and then regulates the downstream signaling pathways such as JAK/STAT3, mediating the genesis and development of multiple leukemia subtypes. Most existing studies suggest that ZNF384 is indeed involved in the process of tumor genesis and development as an important signal molecule (12), however, its correlation with breast cancer has not been reported yet. It remains unclear whether ZNF384 can affect the genesis and development of breast cancer, or serve as a molecular marker for diagnosis and prognosis judgement, and a potential therapeutic target.
Our research group first collected tissue samples from clinical breast cancer patients, and 23 common mutant genes from multiple samples were discovered through WES, including ZNF384, PDE4DI, MAP3K1, POT1 and PCM1 and so on. Further enrichment analysis demonstrated that these genes were mainly enriched in cancer, PI3K/Akt and cAMP signaling pathways. Subsequently, the target genes (ZNF384 and PDE4DIP, with the consistency of over 15%) whose changes in expression levels show high correlations with mutation were screened from the above common genes through analyzing TCGA database by using cBioPortal. On this account, the gene expression and mutation data, as well as clinical information from the TCGA database were analyzed, the results revealed that the ZNF384 expression level in breast cancer tissues with ZNF384 mutation was far higher than that in the non-mutation tissues. Moreover, such gene mutation and high expression displayed significantly positive correlation with patient survival (P<0.05). Finally, gene set enrichment analysis (GSEA) was adopted, which showed that there were cell cycle signaling pathway-and mitosis metaphase signaling pathway-related gene enrichments in tissues with high ZNF384 expression and mutation. Besides, by using the MEM software, it was predicted that these pathophysiological effects mediated by ZNF384 mutation and high expression might be related to its regulation on the expression level and activity of its downstream gene CXCL14.
This paper is the first report on the correlation between ZNF384 gene and the survival of breast cancer patients, which can not only serve as a molecular marker for diagnosis and prognostic prediction, but also a potential therapeutic target.
Conclusions
Our findings suggested that overexpression and mutation of ZNF384 was associated with favorable prognosis in breast cancer patients, which can serve as a molecular marker for the diagnosis and prognostic prediction. Therefore, a thorough study into it is worth the effort.
Acknowledgments
Funding: We gratefully acknowledge
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All of the patients have signed the informed consent forms. This study was approved by the Ethics Committee of Women’s Hospital of Zhejiang University School of Medicine (approval ID: 20170122).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Seely JM, Alhassan T. Screening for breast cancer in 2018-what should we be doing today? Curr Oncol 2018;25:S115-S124. [Crossref] [PubMed]
- Kaklamani VG. Developments in Breast Cancer 2017-2018: New Drugs, New Drug Classes-and the Prospect of More to Come. Oncology (Williston Park) 2017;31:6-7. [PubMed]
- Zheng K, Tan JX, Li F, et al. Clinicopathologic Factors Related to the Histological Tumor Grade of Breast Cancer in Western China: An Epidemiological Multicenter Study of 8619 Female Patients. Transl Oncol 2018;11:1023-33. [Crossref] [PubMed]
- Bao Y, Kwok C, Lee CF. Breast cancer screening behaviors among Chinese women in Mainland China. Nurs Health Sci 2018;20:445-51. [Crossref] [PubMed]
- Li S, Wang X, Yang J, et al. Clinicopathological features and survival of early stage breast cancer in northwest China: A population-based retrospective study of 1287 patients. Thorac Cancer 2018;9:10-8. [Crossref] [PubMed]
- Xing P, Dong H, Liu Q, et al. Impact of persistence on survival of patients with breast cancer treated with endocrine therapy in Northeast China: a prospective study. Oncotarget 2017;8:102499-510. [Crossref] [PubMed]
- Elghoroury EA, ElDine HG, Kamel SA, et al. Evaluation of miRNA-21 and miRNA Let-7 as Prognostic Markers in Patients With Breast Cancer. Clin Breast Cancer 2018;18:e721-e726. [Crossref] [PubMed]
- Shidfar A, Costa FF, Scholtens D, et al. Expression of miR-18a and miR-210 in Normal Breast Tissue as Candidate Biomarkers of Breast Cancer Risk. Cancer Prev Res (Phila) 2017;10:89-97. [Crossref] [PubMed]
- Lefort S, Thuleau A, Kieffer Y, et al. CXCR4 inhibitors could benefit to HER2 but not to triple-negative breast cancer patients. Oncogene 2017;36:1211-22. [Crossref] [PubMed]
- Liu Y, Lv DL, Duan JJ, et al. ALDH1A1 expression correlates with clinicopathologic features and poor prognosis of breast cancer patients: a systematic review and meta-analysis. BMC Cancer 2014;14:444. [Crossref] [PubMed]
- Schmidt MK, van den Broek AJ, Tollenaar RA, et al. Breast Cancer Survival of BRCA1/BRCA2 Mutation Carriers in a Hospital-Based Cohort of Young Women. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Hirabayashi S, Ohki K, Nakabayashi K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 2017;102:118-29. [Crossref] [PubMed]
- Yao L, Cen J, Pan J, et al. TAF15-ZNF384 fusion gene in childhood mixed phenotype acute leukemia. Cancer Genet 2017;211:1-4. [Crossref] [PubMed]
- Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35:W169-75. [Crossref] [PubMed]
- Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251-3. [Crossref] [PubMed]
- Zeng JH, Liang L, He RQ, et al. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget 2017;8:16811-28. [PubMed]
- Sparano JA, Golden AA, Montagna C. Translating the TCGA breast cancer results into clinical practice: searching for therapeutic clues. Oncology (Williston Park) 2013;27:1284, 1286.
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Hou GX, Liu P, Yang J, et al. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One 2017;12:e0174515. [Crossref] [PubMed]
- Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol 2014;15:e279-89. [Crossref] [PubMed]
- Huang J, Wang L, Jiang M, et al. Low BIK outside-inside-out interactive inflammation immune-induced transcription-dependent apoptosis through FUT3-PMM2-SQSTM1-SFN-ZNF384. Immunol Res 2016;64:461-9. [Crossref] [PubMed]


