
Knockdown of RPL34 suppresses osteosarcoma cell proliferation likely through EIF3/FAU signaling pathway
Introduction
Osteosarcoma (OS), a cancer which is derived from early-lineage bone-forming mesenchymal cells, predominantly occurs in young adults and children. It is the most frequently occurring malignant bone tumor (1-4), having an incidence rate of approximately 3/1,000,000 every year (5). Although advances in surgical techniques and the use of neoadjuvant chemotherapy have greatly improved the 5-year survival rate of OS patients, the survival trend has stayed stagnant over the past 20 years (6-8), and most patients die of the metastatic disease (9-11). Therefore, there is an urgent need to explore novel diagnostic and prognostic biomarkers and potential therapeutic targets to improve the prognosis in patients with OS. Our team’s study of OS has shown that highly expressed RPL34 is closely related to a poor prognosis, and its knockdown inhibits OS cells proliferation. Bioinformatics analysis revealed that RPL34 might interact with the subunit of the downstream eukaryotic initiation factor EIF3 (EIF3A, EIF3F, EIF3G), and UBA52, UBC, FAU (12).
Moreover, each gene ontology (GO) category was significantly enriched by the three identified genes in the protein-protein interaction (PPI) network, mainly EIF3A, EIF3F, and EIF3G. Therefore, we hypothesized that knockdown of RPL34 inhibits osteosarcoma cell proliferation probably through EIF3A, EIF3F, and EIF3G. This study aims to explore the effects of the knockdown of RPL34 on osteosarcoma cell proliferation through EIF3/FAU.
RPL34 is a highly evolutionarily conserved protein. However, some recent studies have found the dysregulated expression of RPL34 in several cancers. It was observed that RPL34 expression was significantly up-regulated in pancreatic cancer, and the RPL34 high expression was also found in pancreatic cancer patients’ tumors with lymphatic metastasis, angiolymphatic invasion, and positive pAJCC stage (13). Research on human glioma suggested that the knockdown of RPL34 suppresses the migration and proliferation of glioma cells through the inactivation of the JAK/STAT3 signaling pathway (14). Additionally, RPL34 is an oncogene that mediates the metastasis of esophageal cancer cells via an inactivation of the PI3K/Akt signaling pathway (15). Liu et al. (16) showed that the expression of RPL34 was up-regulated significantly in a gastric cancer cell line, and knockdown of RPL34 significantly blocked cell cycle, promoted cell apoptosis, and inhibited cell proliferation in SGC-7901 cells. Yang et al. (17) reported that RPL34 was highly expressed in non-small cell lung cancer tissues compared with adjacent tissues, and the overexpression of RPL34 significantly promoted the differentiation and proliferation of H1299 cells. The expression of RPL34 in oral squamous cell carcinoma was significantly increased, and knockdown of RPL34 could promote cell apoptosis, block cell proliferation, and metastasis (18). These findings suggest that RPL34 is closely related to malignant cell proliferation.
EIF3, the largest family member of all eIFs, contains at least 13 subunits that are consecutively designated EIF3a to EIF3m (19,20). Members of the eIF3 family play a key role in protein translation. EIF3 first binds to the 40S ribosome and then promotes loading of Met-tRNA/eIF2-GTP ternary complex, and finally forms the 43S preinitiation complex. It also notably aids EIF4 to recruit mRNAs into the complex (21). These observations indicate that EIF3 is a key initiation factor in eukaryotes. In addition, aberrant EIF3 expression is found in many types of human malignancies (22-25).
The gene FAU encodes a fusion protein consisting of the ubiquitin-like protein, Fubi, and the S30 ribosomal protein. The gene consists of four introns and five exons, and a housekeeping gene characterizes the 5’ untranslated region. FAU is a tumor suppressor gene. Several reports have shown that FAU is abnormally expressed in osteosarcoma cells and is involved in the regulation of apoptosis (26-28). Little research has been done on the role of UBC and UBA52 in cancer.
Although there are many reports on the expression and mechanism of RPL34, EIF3, and FAU in cancer, the relationship between RPL34 and EIF3/FAU in osteosarcoma is unclear. In this study, we explored the effects of the knockdown of RPL34 on osteosarcoma cells proliferation through EIF3/FAU, hoping that this might help to identify a new prognosis marker and potential therapeutic target for OS in the future.
Methods
Cell culture
The Saos-2 and hFOB1.19 cell lines were sourced from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). DMEM (Thermo, Massachusetts, US) was used to culture the hFOB 1.19 cell line, while McCoy Y’s 5A (Thermo, Massachusetts, US) was used to culture the Saos-2 cell line. Media were supplemented with a 10% fetal bovine serum (FBS), 0.1 mg/mL streptomycin, and 100 U/mL penicillin. Cells were cultured in 5% CO2 in a humidified chamber and maintained at 37 °C.
qRT-PCR
RNAiso Plus reagents (Takara, Osaka, Japan) were used to isolate RNA from whole cells. cDNA was reverse transcribed with 5× Prime Script RT Master MIX (perfect Real Time; Takara, Osaka, Japan) from 0.5 µg of RNA. A total of 0.2 µg of cDNA was used as a template for qRT-PCR. qRT-PCR was performed in a real-time PCR machine (ABI, CA, USA) using SYBR Premix EX Taq (2×) (ABI, Carlsbad, CA, USA). The PCR cycling parameters were as follows: 40 cycles of 95 °C for 3 min, 95 °C for 10 s, and 60 °C for 30 s. The housekeeping reference gene was GAPDH. The comparative threshold cycle method (△△Ct) was used to determine mRNA expression levels (29). Primers used for PCR (Gene Wiz, Shanghai, China) are described as follows: RPL34, 5'-GTTTGACATACCGACGTAGGC-3'and5'-GCACACATGGAACCACC ATAG-3'; EIF3A, 5'-AGAGTAGAGCGCCTGTACCA-3' and 5'-TGGTTATGGTGGCGCTGAAT-3'; EIF3F, 5'-ACACAAGTCTCCAGAACGGC-3' and 5'-TCCAATCACTCTGTT GGGGC-3'; UBA52, 5'-CTTTTTCTTCAGCGAGGCGG-3' and 5-TCAATGGTGTCACT GGGCTC-3'; UBC, 5'-TCGGCCTTAGAACCCCAGTA-3' and 5'-GAGATCCCTCCGCAG AATCG-3'; FAU, 5'-CCAGGAGCTACACACCTTCG-3' and 5'-GACTTGATCTTCCGGGGCAA-3'; EIF3G, 5'-CAAGATAGATGAGGATGGCAAGA-3' and 5'-GGTCCTCTTTGCTGGTGATGA-3'; GAPDH, 5'-TGACAACTTTGGTATCGTGGAAGG-3' and 5'-AGGCAG GGATGATG TTCTGGAGAG -3'.
Construction of lentiviral vectors and cell transfection
RPL34 mRNA (GenBank accession No. NM_000995.4) was targeted for depletion using short hairpin RNA (shRNA, CACAGAGTCAGAAAGCTAA) targeting a region of the RPL34 cDNA sequence (sh-RPL34) and a negative control shRNA (sh-NC, TTCTCCGAACGTG TCACGT) was used as a control. Gene Wiz Co. Ltd. (Shanghai, P.R. China) designed and synthesized the shRNA constructs. The sh-RNA sequence was inserted into the pLKO.1-PURO Linear GFP-containing vector having the Agel/EcoRI sites (Gene Wiz Co. Ltd. Shanghai, China). This lentiviral shRNA vector was then co-transfected in 293T cells. Saos-2 cells had RPL34-shRNA or control shRNA transfected into them using Lipofectamine 2000 (Invitrogen, CA, USA). A fluorescence microscope (AMI, CA, USA) was used to confirm the efficiency of transfection after 2 days, whereas qRT-PCR and Western blot confirmed knockdown efficiency.
Western blot
The cells were harvested and then lysed on ice for 20 minutes with a pre-cooled lysis buffer (RIPA Lysis Buffer III, Sangon Biotech, Shanghai, China). The lysate was centrifuged at 12,000 rpm at 4 °C for 15 minutes, and then the supernatant was collected. Protein concentration was measured using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Ten percent SDS-PAGE gels were utilized to resolve 30 µg of protein from each sample. The proteins were then transferred onto polyvinylidene difluoride (PVDF) (Millipore, MA, USA) membranes. The membranes were blocked for 1 h with PBST containing 5% non-fat dry milk powder. The PVDF membranes were incubated with rabbit anti-FLAG primary antibody (1:1,000, SAB, Shanghai, China) overnight at 4 °C and then with goat anti-rabbit IgG (H+L) secondary antibody (1:1,000, Proteintech Group, Chicago, USA) for 2 hours at room temperature. The PVDF membranes were soaked in PBST buffer and washed slowly for 10 minutes. The PVDF membranes were placed on the plastic wrap, and then the ECL reagent (Millipore, MA, USA) was added to the PVDF membranes. The enhanced chemiluminescence assay was performed using the ChemiDoc XRS+ (Bio-Rad, Minnesota, USA) according to the manufacturer’s instructions. β-actin was used as a normalization control. Finally, the grayscale value analysis was carried out with ImageJ software.
Screening for genes associated with RPL34 using bioinformatics.
The genes regulated by RPL34 were obtained from the University of California Santa Cruz’s ENCODE Genome Browser (http://genome.ucsc.edu), based on human transcription factor-gene pairs. A high-confidence protein interaction was retrieved from the STRING database to construct a PPI network, and network analysis of proteins interacting with RPL34 was performed (30). The PPI combination score >0.9 was set as the threshold for screening interaction pairs. Finally, Cytoscape software was used to visualize the corresponding network. The genes, identified by the PPI network, were subjected to GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The number of enriched genes ≥2 and the threshold value of P<0.05 were the screening criteria for significant enrichment of GO terms or KEGG pathways (12).
Statistical analysis
Data are presented as mean ± standard deviation, with each experiment conducted three times. The t-test was used for the quantitative comparison between two groups. GraphPad Prism 5.0 (San Diego, CA, USA) or SPSS software, 20.0 (Chicago, IL, USA) were used for all statistical analyses. A P<0.05 was the threshold for statistical significance.
Results
Screening for genes associated with RPL34 using bioinformatics
Our previous research team found that RPL34 may regulate the proliferation of osteosarcoma cells through the subunits of the downstream eukaryotic initiation factors EIF3 (EIF3A, EIF3F, EIF3G) UBA52, UBC, and FAU. Interestingly, each GO category is significantly enriched by EIF3A, EIF3F and EIF3G identified in the PPI network (12). Therefore, it was speculated that knockdown of RPL34 suppresses osteosarcoma cell proliferation through EIF3A, EIF3F, and EIF3G.
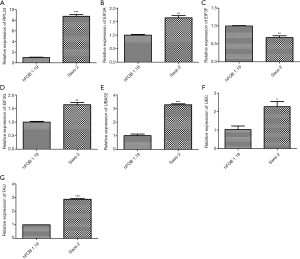
Expression levels of RPL34, EIF3A, EIF3F, and FAU were increased in Saos-2 cells compared to hFOB1.19 cells
qRT-PCR was performed to quantify the expression levels of RPL34, EIF3A, EIF3F, EIF3G, UBA52, UBC, and FAU mRNA in hFOB1.19 cells and Saos-2 OS cells. The mRNA expression levels of RPL34, EIF3A, EIF3G, UBA52, UBC and FAU were significantly elevated in the human Saos-2 cell line relative to the hFOB1.19 cell line (Figure 1, *, P<0.05, **, P<0.01, ***, P<0.001). EIF3F expression was significantly decreased in the Saos-2 cell line (Figure 1C, **, P<0.01).
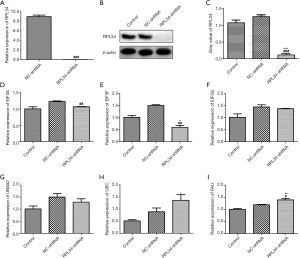
Knockdown of RPL34 suppresses osteosarcoma cells proliferation likely through EIF3/FAU signaling pathway
Our previous research team found that knockdown of RPL34 significantly inhibits MG-63, U2OS, Saos-2, and HOS Cells proliferation (12). In order to explore the effect of knockdown of RPL34 on EIF3A, EIF3F, and FAU expression, we induced shRNA-targeting RPL34 into Saos-2 cells. RT-qPCR and western blot confirmed the knockdown efficiency of RPL34. The observation showed that the expression of RPL34 mRNA was significantly down-regulated in the sh-RPL34 transfected cells compared to the cells transfected with NC-shRNA (Figure 2A, ###, P<0.001). This result was similar to the protein level (Figure 2B,C, ###, P<0.001). The EIF3A and EIF3F mRNA expression levels in Saos-2 cells transfected with sh-RPL34 were significantly down-regulated compared to those transfected with NC-shRNA (Figure 2D,E, ##, P<0.01). There was no significant change in the relative expression of EIF3G and UBA52 genes (Figure 2F,G, *, P>0.05, #, P>0.05). Although the relative expression of UBC gene in the sh-RPL34 group was significantly up-regulated relative to the control group (Figure 2H, *, P<0.05), it was unchanged in the RPL34-shRNA group compared to the NC-shRNA group (Figure 2H, #, P>0.05). The relative expression of the FAU gene in the RPL34-shRNA group was significantly up-regulated compared to the control and NC-shRNA groups (Figure 2I, **, P<0.01, #, P<0.05).
Discussion
RPL34 plays an oncogene role in human tumors, especially in malignant tumor cell proliferation. Although our research team found that highly expressed RPL34 is correlated with a poor prognosis, and knockdown of RPL34 inhibits OS cell proliferation (12), the underlying molecular mechanisms by which RPL34 regulates osteosarcoma cell proliferation remains unclear. The results of the present study have shown that the mRNA expression levels of RPL34, EIF3A, EIF3G, UBA52, UBC, and FAU were significantly increased and EIF3F expression was significantly decreased in Saos-2 cells relative to hFOB1.19 cells. Moreover, knockdown of RPL34 mRNA significantly suppresses EIF3A and EIF3F expression and significantly up-regulated FAU expression in Saos-2 cell lines, while the expression of EIF3G, UBA52, and UBC had no significant difference. Our findings further contribute to the evidence linking the knockdown of RPL34 to the suppression of osteosarcoma cell proliferation likely through EIF3A, EIF3F, and FAU.
EIF3A is a key functional component of ribosomes, and not only promotes the establishment of the ribosomal complex and initiates translation, but also regulates tumor pathogenesis and therapeutic efficacy. As a proto-oncogene, EIF3A can promote tumorigenesis. It is believed that the dysregulation of translation initiation is a major factor in promoting tumorigenesis. Several reports have shown that EIF3A is highly expressed in a range of cancers including the urinary bladder (23), oral cavity (22), and pancreas (24). EIF3A expression is associated with cancer metastasis because high EIF3A expression not only promotes tumor aggressiveness but also enhances the migration and invasion of cancer cells in pancreatic cancer (24).
EIF3F regulates the initiation of translation and is the p47 subunit of the eIF3 multimeric complex (31,32). In human cancers, EIF3F plays a negative regulatory role in translation (33). Dysregulated EIF3F expression has been detected in multiple types of cancer. EIF3F overexpression induces apoptosis in pancreatic and melanoma cancer cells and disrupts the proliferation of tumor cells (32,34,35). Li et al. (36) found that EIF3F expression was elevated in adjacent non-cancerous tissues relative to gastric cancer tissues and that EIF3F levels were correlated with the stage of tumor progression and the probability of recurrence, suggesting that lower levels of EIF3F are predictive of poorer outcomes in gastric cancer patients.
Overexpression of FAU plays a carcinogenic role, promoting anchoring independence and transformation of human osteosarcoma cells (26). Also, several reports have shown that FAU acts as a pro-apoptotic regulatory gene (27,28). Apoptosis is central to many developmental processes including immune response, differentiation, oncogenesis, and degenerative disease, and is a tightly regulated process (37,38). The regulatory role of FAU in the context of apoptosis is a key component of the oncogenic process. Induction of apoptosis is also a mechanism by which many therapeutic agents combat cancers (38,39). More importantly, our data show that knockdown of RPL34 mRNA significantly up-regulates FAU expression, indicating that knockdown of RPL34 suppresses the proliferation of osteosarcoma cells probably through FAU.
Conclusions
In summary, the data we provided further suggests that the knockdown of RPL34 suppresses osteosarcoma cell proliferation likely through EIF3A, EIF3F, and FAU. These findings might help us to explore targeted therapeutic markers and signaling pathways for osteosarcoma treatment in the future.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.05.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was approved by the medical ethics committee of the First Affiliated Hospital of Guangxi Medical University (NO.2017-KY-GJ-055).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gill J, Ahluwalia MK, Geller D, et al. New targets and approaches in osteosarcoma. Pharmacol Ther 2013;137:89-99. [Crossref] [PubMed]
- Wang Q, Shi Z, Zhang H, et al. Upregulation of exosomal integrin β4 causes osteosarcoma cell proliferation via the PI3K-Akt-mTOR signaling pathway. Translational Cancer Research 2018;7:1209-20. [Crossref]
- Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet 2013;45:799-803. [Crossref] [PubMed]
- Yang Z, Yang M, Tao H, et al. Effect of 17β-estradiol on the migration ability of human osteosarcoma U2OS cells. Translational Cancer Research 2018;7:1221-7. [Crossref]
- Errani C, Longhi A, Rossi G, et al. Palliative therapy for osteosarcoma. Expert Rev Anticancer Ther 2011;11:217-27. [Crossref] [PubMed]
- Allison DC, Carney SC, Ahlmann ER, et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma 2012;2012:704872. [Crossref] [PubMed]
- Miller BJ, Cram P, Lynch CF, et al. Risk factors for metastatic disease at presentation with osteosarcoma: an analysis of the SEER database. J Bone Joint Surg Am 2013;95:e89. [Crossref] [PubMed]
- Dong Z, Sun K, Luan YJ, et al. Expression and clinical significance of threonine and tyrosine protein kinase (TTK) in osteosarcoma. Translational Cancer Research 2017;6:285-92. [Crossref]
- Marko TA, Diessner BJ, Spector LG. Prevalence of Metastasis at Diagnosis of Osteosarcoma: An International Comparison. Pediatric Blood & Cancer 2016;63:1006-11. [Crossref] [PubMed]
- Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiology 2015;39:593-9. [Crossref] [PubMed]
- Wu H, Xia LM. siRNA-mediated silencing of FOXQ1 expression inhibits cell proliferation and induces apoptosis in human osteosarcoma cells. Translational Cancer Research 2017;6:1258-62. [Crossref]
- Luo S, Zhao J, Fowdur M, et al. Highly expressed ribosomal protein L34 indicates poor prognosis in osteosarcoma and its knockdown suppresses osteosarcoma proliferation probably through translational control. Sci Rep 2016;6:37690. [Crossref] [PubMed]
- Wei F, Ding L, Wei Z, et al. Ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells. Oncotarget 2016;7:85259-72. [Crossref] [PubMed]
- Ji P, Wang L, Liu J, et al. Knockdown of RPL34 inhibits the proliferation and migration of glioma cells through the inactivation of JAK/STAT3 signaling pathway. J Cell Biochem 2019;120:3259-67. [Crossref] [PubMed]
- Fan H, Li J, Jia Y, et al. Silencing of Ribosomal Protein L34 (RPL34) Inhibits the Proliferation and Invasion of Esophageal Cancer Cells. Oncol Res 2017;25:1061-8. [Crossref] [PubMed]
- Liu H, Liang S, Yang X, et al. RNAi-mediated RPL34 knockdown suppresses the growth of human gastric cancer cells. Oncol Rep 2015;34:2267-72. [Crossref] [PubMed]
- Yang S, Cui J, Yang Y, et al. Over-expressed RPL34 promotes malignant proliferation of non-small cell lung cancer cells. Gene 2016;576:421-8. [Crossref] [PubMed]
- Dai J, Wei W. Influence of the RPL34 gene on the growth and metastasis of oral squamous cell carcinoma cells. Arch Oral Biol 2017;83:40-6. [Crossref] [PubMed]
- Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol Hematol 2006;59:169-80. [Crossref] [PubMed]
- Yin JY, Dong Z, Liu ZQ, et al. Translational control gone awry: a new mechanism of tumorigenesis and novel targets of cancer treatments. Biosci Rep 2011;31:1-15. [Crossref] [PubMed]
- Wagner S, Herrmannova A, Malik R, et al. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol Cell Biol 2014;34:3041-52. [Crossref] [PubMed]
- Spilka R, Laimer K, Bachmann F, et al. Overexpression of eIF3a in Squamous Cell Carcinoma of the Oral Cavity and Its Putative Relation to Chemotherapy Response. J Oncol 2012;2012:901956. [Crossref] [PubMed]
- Spilka R, Ernst C, Bergler H, et al. eIF3a is over-expressed in urinary bladder cancer and influences its phenotype independent of translation initiation. Cell Oncol (Dordr) 2014;37:253-67. [Crossref] [PubMed]
- Wang SQ, Liu Y, Yao MY, et al. Eukaryotic Translation Initiation Factor 3a (eIF3a) Promotes Cell Proliferation and Motility in Pancreatic Cancer. J Korean Med Sci 2016;31:1586-94. [Crossref] [PubMed]
- Li G, Wang K, Li Y, et al. Role of eIF3a in 4-amino-2-trifluoromethyl-phenyl retinate-induced cell differentiation in human chronic myeloid leukemia K562 cells. Gene 2019;683:195-209. [Crossref] [PubMed]
- Rossman TG, Visalli MA, Komissarova EV. fau and its ubiquitin-like domain (FUBI) transforms human osteogenic sarcoma (HOS) cells to anchorage-independence. Oncogene 2003;22:1817-21. [Crossref] [PubMed]
- Pickard MR, Mourtada-Maarabouni M, Williams GT. Candidate tumour suppressor Fau regulates apoptosis in human cells: an essential role for Bcl-G. Biochim Biophys Acta 2011;1812:1146-53. [Crossref] [PubMed]
- Mourtada-Maarabouni M, Kirkham L, Farzaneh F, et al. Regulation of apoptosis by fau revealed by functional expression cloning and antisense expression. Oncogene 2004;23:9419-26. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Masutani M, Sonenberg N, Yokoyama S, et al. Reconstitution reveals the functional core of mammalian eIF3. EMBO J 2007;26:3373-83. [Crossref] [PubMed]
- Shi J, Kahle A, Hershey JW, et al. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 2006;25:4923-36. [Crossref] [PubMed]
- Doldan A, Chandramouli A, Shanas R, et al. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog 2008;47:235-44. [Crossref] [PubMed]
- Marchione R, Laurin D, Liguori L, et al. MD11-mediated delivery of recombinant eIF3f induces melanoma and colorectal carcinoma cell death. Mol Ther Methods Clin Dev 2015;2:14056. [Crossref] [PubMed]
- Hershey JW. Regulation of protein synthesis and the role of eIF3 in cancer. Braz J Med Biol Res 2010;43:920-30. [Crossref] [PubMed]
- Li G, Wang N, Sun C, et al. Decreased expression of eukaryotic initiation factor 3f is an adverse prognostic factor for stage I-III gastric cancer. World J Surg Oncol 2014;12:72. [Crossref] [PubMed]
- Green DR, Evan GI. A matter of life and death. Cancer Cell 2002;1:19-30. [Crossref] [PubMed]
- Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 2002;109:S97-107. [Crossref] [PubMed]
- Ferreira CG, Epping M, Kruyt FA, et al. Apoptosis: target of cancer therapy. Clin Cancer Res 2002;8:2024-34. [PubMed]


