
Competitive endogenous RNA network identifies four long non-coding RNA signature as a candidate prognostic biomarker for lung adenocarcinoma
Introduction
Lung cancer (LC) is among the most common types of malignancies and is the first leading cause of cancer-related death worldwide (1). In recent years, the morbidity and mortality of LC have been shown an increasing tendency, especially in some regions with higher tobacco consumption (2). According to pathological type, LC mainly includes small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), and NSCLC is the major pathological type and accounts for approximately 85% of LC (3). Although some significant advances were obtained in new detecting and treating methods, the 5-year overall survival (OS) rate for all stages of NSCLC less than 20% (4). The main causes lie in that only approximately 15% NSCLC patients are timely determined at an early stage (I or II stage) and NSCLC tends to form metastasis at an early stage (5). If the majority of NSCLC patients are timely diagnosed at an early stage, active treatments of NSCLC are able to offer a favorable prognosis and the 5-year OS rate will raise to 70–90% (6).
Among NSCLC, lung adenocarcinoma (LUAD) is the major histological subtype and annually results in more than 600,000 deaths worldwide (7). Currently, the diagnosis and prognosis of LUAD is evaluated on the basis of histological grade, disease stage and the expression level of some key genes. However, due to the heterogeneity of LUAD, the clinical outcomes are highly variable and the predictive values are limited on the basis of clinical and pathological characteristics in detecting LUAD at an early stage (8). Even though some progresses have been obtained in pathological mechanisms involved in LUAD (8), molecular mechanisms underlying LUAD remain vague. Thus, it is vital to identify potentially molecular diagnostic and prognostic biomarkers and/or therapeutic targets for combating LUAD.
Non-coding RNAs (ncRNAs) are no-coding parts of genome at different cell status and account for more than 90% RNA transcripts (9). Recently, more and more studies have revealed the important regulatory roles of ncRNAs in tumor biology involving in formation, metastasis, prognosis and recurrence of tumor (10). Long non-coding RNAs (lncRNAs), an important class of ncRNA with over 200 nucleotides long (11), are widely reported to play as important regulatory components in tumor promoter and suppressor pathways (10,12). However, little knowledge is known about the roles of lncRNAs in tumor biology. Nevertheless, due to high tissue specificity of lncRNA expression, lncRNAs are still often considered as potentially valuable diagnostic biomarkers of various malignant tumors (13-16). Therefore, identifying LUAD-specific lncRNAs as biomarkers is of clinical value for diagnosis and prognosis of LUAD. Recently, several studies have reported that some lncRNAs were associated with formation and progression of LUAD (17-19), and some were predicted as biomarkers related with clinical outcomes (20,21). But, owing to limited tissue samples, the results from different studies are inconsistent, and lncRNA-related studies with large tissue samples are urgently needed to identify key lncRNAs associated with LUAD. The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) is a publicly available database which can provide large-scale multi-dimensional cancer-related molecular profile data. The current study used LUAD-related data with large sample size from TCGA database to identify LUAD-specific lncRNAs, and increase statistical reliability of this study.
The competitive endogenous RNA (ceRNA) hypothesis was proposed as a novel regulatory mechanism between coding messenger RNAs (mRNAs) and ncRNAs (22). Studies have demonstrated that lncRNAs as ceRNAs competitively combine with microRNAs (miRNAs) by miRNA-response-elements (MREs) to play the key roles in various biological processes (BP) such as tumor formation and metastasis (23). For instance, Liu et al. have proved that two lncRNAs DLEU2 and DDX11-AS1 in lncRNA-miRNA-mRNA ceRNA network were significantly up-regulated in gastric cancer (GC) tissues, and promoted GC cell proliferation by acting as has-miR-30-5p and has-miR-145-5p sponge (24). In serous ovarian cancer, lncRNA PTAR could promote epithelial-to-mesenchymal transition (EMT) and invasion-metastasis by competitively binding miR-101-3p to regulate ZEB1 expression (25). Fan et al. found that the aberrant expression of 4-lncRNA signature (ADAMTS9-AS1, LINC00536, AL391421.1 and LINC00491) in ceRNA network played the key roles in the progression and prognosis of breast cancer (26). In addition, a study has revealed that 7-lncRNA signature in ceRNA network was linked to clinical features in rectal adenocarcinoma (27). In summary, these studies manifest that the dysregulation of lncRNAs in ceRNA network can regulate lncRNA and mRNA interaction by miRNA mediating, and further contribute to the initiation and progression of cancer (28). However, although a few studies have paid attention to LC ceRNAs (29,30), little information is available for LC ceRNAs, especially LUAD ceRNAs.
In the present study, LUAD-related RNA sequencing (RNA-seq) data were retrieved from TCGA database. lncRNA-related ceRNA network was established and LUAD-specific lncRNAs were identified. Further, survival analysis of key lncRNAs was performed to predict important lncRNAs associated with OS of LUAD patients. The present study provides some novel insights into the regulatory mechanism of lncRNAs though ceRNA network in LUAD, and identifying lncRNA signature may serve as novel potentially diagnostic biomarkers and/or therapeutic targets.
Methods
RNA-seq data and patient information collection
RNA-seq data associated with LUAD were retrieved from TCGA database portal (https://cancergenome.nih.gov/). The exclusion criteria of LUAD-related tissues RNA-seq data were included as follows: (I) histological diagnosis for not LUAD; (II) except LUAD suffering from other malignancy; (III) data with incomplete clinical information. Finally, a total of 535 LUAD-related and 59 non-LUAD lung tissues RNA-seq data were retrieved. The clinical information of all LUAD samples patients were simultaneously downloaded from TCGA database. Similarly, LUAD-related miRNA RNA-seq data and clinical information in this study were downloaded from TCGA database. Finally, a total 521 LUAD-related and 46 non-LUAD lung tissues RNA-seq data was included. All RNA-seq data and clinical information were obtained from public TCGA database, and were approved by the Institutional Review Board of the relevant participating institutions and by the National Cancer Institute of NIH. No approval from the ethics committee was required. The present study meets the requirements of data usage and publishing from TCGA database.
RNA-seq data preprocessing and differentially expressed analysis
All raw RNA-seq data were subjected to normalization using the trimmed mean of M-values method.
EdgeR software package in Bioconductor project (version 3.8, http://www.bioconductor.org/) based on R (version 3.5.1, https://www.r-project.org/) language environment was used to screen the differentially expressed lncRNAs (DElncRNAs), differentially expressed mRNAs (DEmRNAs) and differentially expressed miRNAs (DEmiRNAs) between LUAD tissues and non-LUAD lung tissues samples (31). |Log fold change (logFC)| >2 and P<0.01 were set as cut-off criteria. The volcano plots of RNA expression were visualized using the ggplot2 (version 3.1.0, https://github.com/tidyverse/ggplot2) software package. The heat map of differentially expressed RNAs (DERNAs) was plotted using the pheatmap (version 1.0.10) software package.
Construction of lncRNA-miRNA-mRNA ceRNA network
The lncRNA-miRNA-mRNA ceRNA network was established based on ceRNA hypothesis that lncRNA directly combine miRNA by acting as miRNA sponge to indirectly regulate the function of mRNA (32). The ceRNA network was constructed according to the following steps: (I) three types of RNAs with |logFC| >2 and P<0.01 cut-off criteria were kept; (II) the potential target miRNAs of DElncRNAs, as well as the interaction relationships between DElncRNAs and target miRNAs were predicted using the online miRcode tool (miRcode 11, http://www.mircode.org/); (III) the potential target mRNAs of DEmiRNAs were predicted using the online tools including TargetScan (release 7.2, http://www.targetscan.org/), miRDB (version 5.0, http://mirdb.org/) and miRTarBase (release 7.0, http://mirtarbase.mbc.nctu.edu.tw/); (IV) based on ceRNA hypothesis, intersecting miRNAs negatively regulated by lncRNAs and mRNAs were selected to construct ceRNA network. The lncRNA-miRNA-mRNA ceRNA network was constructed and visualized using an open-source Cytoscape (version 3.7.0, https://cytoscape.org/) software (33).
Construction of protein-protein interaction network and analysis
The interactive relationships among DEmRNAs encoding proteins in ceRNA network was analyzed by constructing a protein and protein interaction (PPI) network. The interactive information among DEmRNAs encoding proteins was retrieved from online STRING database (version 11.0, https://string-db.org/) (34). The gene pairs with a combined score ≥0.7 were used to construct the PPI network. The Cytoscape software was used to construct and visualize the interactive relationships among genes within PPI network (version 3.7.0, http://www.cytoscape.org/) (33). The subnetwork was extracted from whole PPI network using Molecular COmplex DEtection (MCODE) algorithm based on topological properties of PPI network, and a plugin MCODE (version 1.5.1) in Cytoscape was used to perform MCODE analysis (35). The threshold parameters were set for degree cutoff =2, node score cutoff =0.2, K-core =2 and max-depth =100.
Identification of key genes
Key genes in PPI network were identified using Centrality analysis. Centrality analyses including subgraph centrality, degree centrality, eigenvector centrality, betweenness centrality, network centrality, information centrality and closeness centrality were performed using a plugin CytoNCA (version 2.1.6) in Cytoscape (36). The genes with higher centrality scores were identified as key gene, and essential genes were identified as intersecting genes of key genes obtained by seven centrality methods. GEPIA database (http://gepia.cancer-pku.cn) is an interactive web server for analyzing gene expression data of tumors and normal tissues from TCGA and genotype-tissue expression database (37), and was used to validate essential genes in PPI network.
LUAD-specific prognostic lncRNA signatures identification
The associations between DElncRNAs in ceRNA network and OS in LUAD patients were evaluated using Kaplan-Meier (KM) estimate and log-rank (LR) test in survival (version 2.43-3) package based on R. The statistical P<0.01 was considered as the significant association between lncRNA and OS. Univariate Cox proportional hazards regression model was performed to evaluate the association between DElncRNAs in ceRNA network and OS. Subsequently, multivariate Cox hazards regression model was implemented to assess the prognostic factors for LUAD. The hazards model was established as follows:
Risk socre = ExplncRNA1 × CoelncRNA1 + ExplncRNA2 × CoelncRNA2 + ExplncRNA3 × CoelncRNA3 + ExplncRNAn × CoelncRNAn
Where “Exp” denotes the expression level of lncRNA, and “Coe” is the regression coefficient from the multivariate Cox regression model (38). According to the median of above risk scores, LUAD patients were divided into high-risk and low-risk groups. Receiver operating characteristic (ROC) curve was constructed using survival ROC (version 1.0.3) package based on R, and was used to measure the risk prediction rate of lncRNAs between high- and low-risk groups.
All statistical analyses were performed using R and SPSS softwares.
Results
Identification of DElncRNA, DEmRNA and DEmiRNA
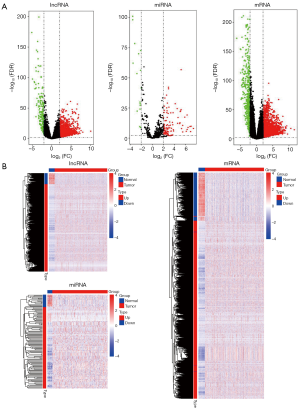
All raw data were normalized using edgeR package, and 8,896 lncRNAs, 18,123 mRNAs, and 672 miRNAs were obtained. Using differentially expressed analysis based on edgeR package, we identified DElncRNAs, DEmRNAs and DEmiRNAs between LUAD tissues samples and non-LUAD lung tissues samples. According to the |logFC| >2 and P<0.01 cut-off criteria, a total of 1,664 DElncRNAs (1,447 up- and 187 down-regulated; http://fp.amegroups.cn/cms/tcr.2019.06.09-1.pdf), 2,503 DEmRNAs (1,975 up- and 528 down-regulated; http://fp.amegroups.cn/cms/tcr.2019.06.09-2.pdf), and 120 DEmiRNAs (104 up- and 16 down-regulated; http://fp.amegroups.cn/cms/tcr.2019.06.09-3.pdf) was identified. The distributions of DElncRNAs, DEmRNAs and DEmiRNAs were displayed using volcano plots in Figure 1A. The expression levels of DElncRNAs, DEmRNAs and DEmiRNAs between LUAD tissues and non-LUAD lung tissues were displayed using heat map in Figure 1B.
Construction of ceRNA network
The relationships among DElncRNAs, DEmiRNAs and DEmRNAs needed to be obtained before lncRNA-miRNA-mRNA ceRNA network was constructed. Using online miRcode tool, the relationships among 1,664 DElncRNAs and 120 DEmiRNAs were evaluated, and finally 140 LUAD-specific DElncRNAs (123 up- and 17 down-regulated) (Table 1) and 33 LUAD-specific DEmiRNAs (28 up- and 5 down-regulated) (Table 2) were putatively identified to interact. Using online tools including TargetScan, miRDB and miRTarBase, we predicted the relationships among DEmiRNAs and their target DEmRNAs. Finally, 57 LUAD-specific target DEmRNAs (41 up- and 16 down-regulated) (Table 3) were putatively identified to interact with 33 LUAD-specific DEmiRNAs.
Table 1
| lncRNA | logFC | P value | FDR |
|---|---|---|---|
| DSCAM-AS1 | 8.035038099 | 5.27E-12 | 2.96E-11 |
| AL160271.1 | 6.952910725 | 4.67E-10 | 2.01E-09 |
| HOTAIR | 6.838380968 | 6.50E-20 | 9.36E-19 |
| DSCR8 | 6.590103737 | 1.61E-11 | 8.52E-11 |
| AC061975.6 | 6.584106301 | 2.85E-21 | 4.63E-20 |
| CLDN10-AS1 | 6.542779442 | 1.12E-30 | 3.81E-29 |
| POU6F2-AS2 | 6.215257277 | 1.55E-14 | 1.20E-13 |
| RMRP | 5.96235355 | 3.45E-08 | 1.16E-07 |
| DSCR4 | 5.930186973 | 7.86E-10 | 3.28E-09 |
| NOVA1-AS1 | 5.885372737 | 1.06E-16 | 1.07E-15 |
| AC020907.1 | 5.854117988 | 2.29E-46 | 1.92E-44 |
| LINC00392 | 5.792399193 | 1.35E-08 | 4.77E-08 |
| UCA1 | 5.77024725 | 2.19E-21 | 3.58E-20 |
| ERVMER61-1 | 5.709669002 | 2.88E-10 | 1.28E-09 |
| LINC00491 | 5.676348335 | 2.20E-15 | 1.88E-14 |
| LINC00501 | 5.327374707 | 5.04E-18 | 5.94E-17 |
| LINC00221 | 5.264633116 | 5.62E-10 | 2.39E-09 |
| AL513123.1 | 5.187897115 | 1.61E-15 | 1.41E-14 |
| NAALADL2-AS2 | 5.123555517 | 5.97E-17 | 6.18E-16 |
| MIR137HG | 5.00623439 | 5.37E-15 | 4.39E-14 |
| ERVH48-1 | 4.849977395 | 2.04E-16 | 1.97E-15 |
| LINC00393 | 4.838707907 | 2.31E-09 | 9.04E-09 |
| AL356133.2 | 4.722511308 | 4.59E-10 | 1.98E-09 |
| LINC01297 | 4.678324477 | 5.36E-11 | 2.64E-10 |
| DLX6-AS1 | 4.552417105 | 1.14E-16 | 1.14E-15 |
| LINC00460 | 4.517552529 | 2.50E-19 | 3.36E-18 |
| LINC00518 | 4.492283543 | 1.06E-13 | 7.45E-13 |
| LINC00355 | 4.434622027 | 3.56E-10 | 1.56E-09 |
| LINC00466 | 4.312624625 | 1.63E-16 | 1.59E-15 |
| POU6F2-AS1 | 4.173659213 | 2.57E-12 | 1.52E-11 |
| LINC00461 | 4.106325477 | 4.03E-24 | 8.10E-23 |
| AC114489.1 | 4.056640519 | 3.35E-15 | 2.81E-14 |
| AC087269.1 | 3.889373143 | 2.52E-22 | 4.44E-21 |
| AC084262.1 | 3.796563805 | 8.76E-19 | 1.11E-17 |
| LINC00473 | 3.73070439 | 4.89E-08 | 1.61E-07 |
| AC010145.1 | 3.711796723 | 1.25E-05 | 2.89E-05 |
| MYCNOS | 3.707588904 | 4.64E-12 | 2.63E-11 |
| LINC00160 | 3.628931995 | 2.65E-21 | 4.31E-20 |
| HOTTIP | 3.608837149 | 8.48E-08 | 2.70E-07 |
| AC080129.1 | 3.505173815 | 1.94E-09 | 7.66E-09 |
| H19 | 3.48727726 | 1.03E-11 | 5.59E-11 |
| LINC00525 | 3.477334167 | 3.60E-24 | 7.27E-23 |
| AC006372.1 | 3.475636767 | 2.89E-07 | 8.46E-07 |
| WASIR2 | 3.466830434 | 2.28E-36 | 1.10E-34 |
| LINC00524 | 3.452370842 | 5.54E-11 | 2.72E-10 |
| LINC00200 | 3.418837644 | 3.91E-05 | 8.37E-05 |
| AC022148.1 | 3.410100434 | 1.08E-13 | 7.56E-13 |
| KIF25-AS1 | 3.402900089 | 9.41E-11 | 4.47E-10 |
| LINC00536 | 3.323145605 | 9.46E-12 | 5.16E-11 |
| SAMSN1-AS1 | 3.238625796 | 8.49E-12 | 4.66E-11 |
| FER1L6-AS1 | 3.230540994 | 3.52E-05 | 7.58E-05 |
| ABCA9-AS1 | 3.229175407 | 2.95E-10 | 1.31E-09 |
| LINC00308 | 3.216837192 | 1.20E-07 | 3.73E-07 |
| STEAP2-AS1 | 3.187740874 | 1.05E-19 | 1.47E-18 |
| AC026320.1 | 3.168057859 | 2.20E-06 | 5.76E-06 |
| LINC00470 | 3.164033391 | 1.16E-08 | 4.13E-08 |
| C20orf197 | 3.118216682 | 2.62E-19 | 3.51E-18 |
| LSAMP-AS1 | 3.078327487 | 3.71E-08 | 1.24E-07 |
| AL354707.1 | 3.049451724 | 2.38E-30 | 7.89E-29 |
| GRM7-AS3 | 3.03536705 | 6.86E-07 | 1.91E-06 |
| C2orf48 | 3.028751823 | 1.74E-29 | 5.39E-28 |
| LINC00488 | 3.002795098 | 4.02E-05 | 8.58E-05 |
| FNDC1-IT1 | 2.998468616 | 8.87E-15 | 7.11E-14 |
| AP002478.1 | 2.966518081 | 5.22E-10 | 2.23E-09 |
| LINC00051 | 2.957193174 | 1.20E-06 | 3.25E-06 |
| LINC00337 | 2.919322952 | 1.42E-22 | 2.59E-21 |
| AC112721.1 | 2.916591282 | 2.70E-15 | 2.28E-14 |
| CHODL-AS1 | 2.908639764 | 3.41E-07 | 9.89E-07 |
| CACNA1C-IT3 | 2.896380101 | 3.04E-05 | 6.63E-05 |
| TDRG1 | 2.816409777 | 6.26E-06 | 1.52E-05 |
| E2F3-IT1 | 2.782045585 | 1.08E-06 | 2.93E-06 |
| AL139002.1 | 2.779424865 | 4.45E-04 | 8.11E-04 |
| AL021395.1 | 2.743868621 | 7.76E-05 | 1.59E-04 |
| PVT1 | 2.730465693 | 4.86E-51 | 5.08E-49 |
| HNF1A-AS1 | 2.706225543 | 8.41E-11 | 4.02E-10 |
| TBL1XR1-AS1 | 2.69669369 | 1.40E-09 | 5.66E-09 |
| LINC00319 | 2.684889139 | 7.97E-10 | 3.32E-09 |
| AC011483.1 | 2.649346027 | 4.89E-06 | 1.20E-05 |
| IGF2-AS | 2.648003097 | 1.12E-06 | 3.03E-06 |
| DPYD-AS2 | 2.597407768 | 2.02E-05 | 4.52E-05 |
| DSCR10 | 2.577147493 | 5.32E-05 | 1.12E-04 |
| LINC00440 | 2.545355975 | 6.39E-05 | 1.33E-04 |
| LINC00519 | 2.499562758 | 1.37E-16 | 1.36E-15 |
| VCAN-AS1 | 2.495803444 | 8.64E-09 | 3.14E-08 |
| LPP-AS1 | 2.471177962 | 3.18E-04 | 5.93E-04 |
| AL353803.1 | 2.457852946 | 3.99E-09 | 1.51E-08 |
| CHL1-AS1 | 2.433177056 | 4.30E-11 | 2.14E-10 |
| IL20RB-AS1 | 2.431801592 | 3.68E-07 | 1.06E-06 |
| EGOT | 2.39267871 | 9.09E-16 | 8.17E-15 |
| ARHGEF3-AS1 | 2.391983831 | 1.25E-04 | 2.49E-04 |
| C11orf44 | 2.374776206 | 1.24E-07 | 3.85E-07 |
| ATG10-AS1 | 2.3672828 | 3.11E-04 | 5.80E-04 |
| SOX21-AS1 | 2.341296999 | 8.79E-11 | 4.19E-10 |
| U52111.1 | 2.324981608 | 7.29E-27 | 1.89E-25 |
| AC007731.1 | 2.313251661 | 2.04E-05 | 4.56E-05 |
| GRM5-AS1 | 2.313149986 | 4.62E-09 | 1.73E-08 |
| AC012640.1 | 2.290965585 | 1.11E-17 | 1.24E-16 |
| AL117190.1 | 2.26875888 | 3.03E-08 | 1.02E-07 |
| FOXP1-IT1 | 2.265388943 | 3.12E-07 | 9.10E-07 |
| C1orf220 | 2.256853758 | 1.93E-41 | 1.30E-39 |
| AC092535.1 | 2.240514584 | 3.66E-13 | 2.42E-12 |
| LINC00485 | 2.213870708 | 6.88E-05 | 1.42E-04 |
| AL391152.1 | 2.211300987 | 2.47E-08 | 8.45E-08 |
| LINC00330 | 2.206368932 | 2.54E-09 | 9.89E-09 |
| AL139385.1 | 2.157764324 | 9.98E-14 | 7.04E-13 |
| AC009121.1 | 2.147340838 | 5.76E-21 | 9.09E-20 |
| SYNPR-AS1 | 2.124398098 | 6.37E-14 | 4.60E-13 |
| MEG3 | 2.109729516 | 8.26E-11 | 3.95E-10 |
| ZBTB20-AS3 | 2.109085084 | 3.37E-03 | 5.38E-03 |
| AC110921.1 | 2.09676636 | 1.19E-04 | 2.38E-04 |
| ANO1-AS2 | 2.093601221 | 1.73E-05 | 3.91E-05 |
| C10orf91 | 2.088023604 | 4.44E-14 | 3.28E-13 |
| LINC00184 | 2.084679983 | 1.93E-11 | 1.01E-10 |
| ARHGAP26-AS1 | 2.08038906 | 5.42E-08 | 1.77E-07 |
| AP000525.1 | 2.07904602 | 1.64E-12 | 9.97E-12 |
| AC110619.1 | 2.078737722 | 5.08E-13 | 3.29E-12 |
| AC016773.1 | 2.072668012 | 1.41E-32 | 5.52E-31 |
| HECW1-IT1 | 2.055407306 | 1.00E-03 | 1.74E-03 |
| AL365356.1 | 2.054075041 | 9.92E-08 | 3.14E-07 |
| LINC00342 | 2.046933782 | 2.23E-14 | 1.69E-13 |
| LNX1-AS2 | 2.038040167 | 3.66E-13 | 2.42E-12 |
| ST7-OT4 | 2.029954915 | 5.47E-11 | 2.69E-10 |
| AL136307.1 | 2.024808806 | 1.16E-10 | 5.44E-10 |
| C5orf17 | 2.013111112 | 8.65E-05 | 1.76E-04 |
| AGAP11 | −2.011064057 | 1.31E-33 | 5.41E-32 |
| AC025431.1 | −2.042390692 | 3.47E-14 | 2.60E-13 |
| RMST | −2.043478157 | 2.23E-16 | 2.14E-15 |
| C5orf64 | −2.059870307 | 4.06E-39 | 2.37E-37 |
| TTTY16 | −2.084328481 | 9.30E-09 | 3.37E-08 |
| LINC00472 | −2.112855406 | 7.57E-42 | 5.26E-40 |
| AC004832.1 | −2.157836408 | 6.01E-21 | 9.44E-20 |
| MED4-AS1 | −2.230027871 | 1.71E-60 | 2.58E-58 |
| LINC00211 | −2.236690384 | 1.13E-34 | 5.00E-33 |
| SRGAP3-AS2 | −2.264937584 | 6.69E-14 | 4.81E-13 |
| MYO16-AS1 | −2.301571571 | 4.46E-16 | 4.15E-15 |
| AP003064.2 | −2.419574318 | 6.46E-26 | 1.52E-24 |
| ADAMTS9-AS1 | −2.702606823 | 7.95E-72 | 2.08E-69 |
| NAV2-AS2 | −2.725182168 | 3.86E-42 | 2.81E-40 |
| AC105206.1 | −2.919107183 | 3.89E-26 | 9.40E-25 |
| AP000438.1 | −2.931393823 | 2.16E-69 | 4.94E-67 |
| LINC00163 | −3.360907124 | 2.56E-76 | 7.86E-74 |
lncRNAs, long non-coding RNAs; ceRNA, competitive endogenous RNA; LUAD, lung adenocarcinoma; logFC, log fold change; FDR, false discovery rate.
Table 2
| miRNA | logFC | P value | FDR |
|---|---|---|---|
| hsa-mir-372 | 7.437105993 | 2.82E-09 | 1.14E-08 |
| hsa-mir-371a | 6.930230254 | 1.48E-06 | 4.17E-06 |
| hsa-mir-122 | 6.145811708 | 8.03E-07 | 2.36E-06 |
| hsa-mir-373 | 5.499517437 | 3.07E-06 | 8.33E-06 |
| hsa-mir-210 | 5.210272814 | 1.86E-59 | 1.56E-57 |
| hsa-mir-31 | 4.647968248 | 1.09E-18 | 9.30E-18 |
| hsa-mir-137 | 4.58286988 | 8.45E-14 | 5.62E-13 |
| hsa-mir-301b | 3.758486051 | 6.94E-23 | 8.80E-22 |
| hsa-mir-196b | 3.636060212 | 1.24E-14 | 8.52E-14 |
| hsa-mir-520c | 3.513854766 | 2.37E-03 | 4.22E-03 |
| hsa-mir-449b | 3.379333024 | 7.21E-07 | 2.13E-06 |
| hsa-mir-215 | 3.16511543 | 6.43E-10 | 2.86E-09 |
| hsa-mir-539 | 3.14438329 | 6.45E-15 | 4.51E-14 |
| hsa-mir-135b | 3.00798826 | 1.30E-27 | 2.24E-26 |
| hsa-mir-192 | 3.001749646 | 1.06E-11 | 5.52E-11 |
| hsa-mir-205 | 2.979998846 | 6.62E-10 | 2.93E-09 |
| hsa-mir-96 | 2.921890226 | 1.21E-44 | 5.81E-43 |
| hsa-mir-301a | 2.729130222 | 4.33E-31 | 9.10E-30 |
| hsa-mir-503 | 2.669107842 | 1.62E-24 | 2.37E-23 |
| hsa-mir-216b | 2.644328582 | 3.79E-06 | 1.02E-05 |
| hsa-mir-489 | 2.634622585 | 5.66E-08 | 1.88E-07 |
| hsa-mir-183 | 2.579080091 | 7.78E-38 | 2.18E-36 |
| hsa-mir-187 | 2.573816898 | 5.98E-12 | 3.14E-11 |
| hsa-mir-224 | 2.299109898 | 5.20E-16 | 3.88E-15 |
| hsa-mir-182 | 2.27499865 | 2.09E-35 | 5.40E-34 |
| hsa-mir-449a | 2.194422015 | 1.37E-05 | 3.42E-05 |
| hsa-mir-200a | 2.170578484 | 1.65E-28 | 3.01E-27 |
| hsa-mir-142 | 2.16966105 | 5.66E-25 | 8.45E-24 |
| hsa-mir-195 | –2.082399445 | 1.13E-75 | 1.91E-73 |
| hsa-mir-133b | –2.215563241 | 5.47E-26 | 8.54E-25 |
| hsa-mir-143 | –2.474794351 | 7.37E-73 | 9.90E-71 |
| hsa-mir-184 | –2.73388336 | 2.84E-24 | 3.98E-23 |
| hsa-mir-144 | –3.176354049 | 1.33E-81 | 2.99E-79 |
miRNAs, microRNAs; ceRNA, competitive endogenous RNA; LUAD, lung adenocarcinoma; logFC, log fold change; FDR, false discovery rate.
Table 3
| mRNA | logFC | P value | FDR |
|---|---|---|---|
| HOXC13 | 6.969775966 | 1.34E-22 | 1.04E-21 |
| SALL1 | 5.819710205 | 5.47E-17 | 2.79E-16 |
| HMGA2 | 5.788539651 | 6.22E-23 | 4.96E-22 |
| SALL3 | 5.50017667 | 4.58E-14 | 1.85E-13 |
| FOXL2 | 4.391302919 | 5.25E-12 | 1.80E-11 |
| HOXA10 | 4.22541626 | 4.45E-22 | 3.34E-21 |
| IGF2BP3 | 4.039045069 | 1.64E-26 | 1.63E-25 |
| MYCN | 4.023455847 | 3.76E-18 | 2.11E-17 |
| DEPDC1 | 3.875064177 | 7.07E-55 | 2.62E-53 |
| SMOC1 | 3.851578922 | 6.80E-18 | 3.74E-17 |
| NPTX1 | 3.756848033 | 4.84E-16 | 2.29E-15 |
| COCH | 3.752741591 | 1.55E-28 | 1.75E-27 |
| PSAT1 | 3.525424513 | 3.56E-48 | 1.01E-46 |
| SSX5 | 3.488914604 | 3.94E-05 | 7.16E-05 |
| ELAVL2 | 3.471819269 | 1.08E-15 | 5.00E-15 |
| PBK | 3.428769291 | 2.82E-43 | 6.41E-42 |
| CCNE1 | 3.316197711 | 1.40E-42 | 3.10E-41 |
| CEP55 | 3.288449477 | 1.57E-55 | 6.00E-54 |
| SLC7A11 | 3.095271245 | 2.31E-23 | 1.90E-22 |
| EFNA3 | 3.087555365 | 2.45E-55 | 9.29E-54 |
| EGLN3 | 3.072798412 | 3.48E-28 | 3.87E-27 |
| CCNB1 | 3.009646799 | 9.48E-55 | 3.49E-53 |
| RET | 2.971273555 | 1.82E-13 | 7.01E-13 |
| COL1A1 | 2.912906867 | 8.02E-32 | 1.08E-30 |
| HNF4A | 2.897133691 | 3.35E-09 | 8.92E-09 |
| E2F7 | 2.763285256 | 4.59E-31 | 5.89E-30 |
| CLSPN | 2.756669421 | 6.29E-40 | 1.25E-38 |
| HOXC8 | 2.73647694 | 1.10E-13 | 4.32E-13 |
| TBX18 | 2.694974307 | 2.26E-10 | 6.70E-10 |
| KCNQ5 | 2.649306296 | 4.37E-20 | 2.82E-19 |
| KIF23 | 2.618363931 | 9.18E-43 | 2.04E-41 |
| CBX2 | 2.522364283 | 2.04E-25 | 1.90E-24 |
| CDC25A | 2.380550406 | 2.11E-37 | 3.73E-36 |
| HMGA1 | 2.312774235 | 2.06E-35 | 3.35E-34 |
| CHEK1 | 2.266437974 | 2.31E-43 | 5.28E-42 |
| MCM4 | 2.244689364 | 5.56E-47 | 1.48E-45 |
| COL5A2 | 2.202086917 | 5.60E-28 | 6.12E-27 |
| PFKP | 2.172781043 | 2.30E-33 | 3.36E-32 |
| HOXB7 | 2.110281006 | 1.22E-17 | 6.57E-17 |
| CCNE2 | 2.063965558 | 1.01E-32 | 1.43E-31 |
| MIXL1 | 2.011494845 | 4.66E-17 | 2.39E-16 |
| PTX3 | −2.013354747 | 2.53E-28 | 2.83E-27 |
| PROK2 | −2.017623563 | 1.85E-24 | 1.63E-23 |
| LDLR | −2.086188772 | 7.70E-85 | 7.54E-83 |
| SLC1A1 | −2.170376032 | 8.25E-51 | 2.59E-49 |
| OSCAR | −2.200662297 | 2.40E-67 | 1.48E-65 |
| DLC1 | −2.218474362 | 4.29E-63 | 2.21E-61 |
| KLF4 | −2.25312747 | 4.90E-62 | 2.47E-60 |
| BDNF | −2.310247479 | 1.86E-32 | 2.58E-31 |
| TGFBR3 | −2.529157615 | 2.39E-87 | 2.49E-85 |
| ARHGEF26 | −2.636625493 | 1.18E-125 | 3.39E-123 |
| SELE | −2.909462573 | 9.52E-64 | 5.18E-62 |
| ARC | −3.418597871 | 8.81E-94 | 1.05E-91 |
| CLIC5 | −3.605080302 | 1.00E-118 | 2.42E-116 |
| RS1 | −3.705828725 | 3.10E-95 | 3.82E-93 |
| TMEM100 | −4.315116104 | 7.43E-144 | 3.28E-141 |
| SERTM1 | −4.74437604 | 2.27E-70 | 1.53E-68 |
mRNAs, messenger RNAs; ceRNA, competitive endogenous RNA; LUAD, lung adenocarcinoma; logFC, log fold change; FDR, false discovery rate.
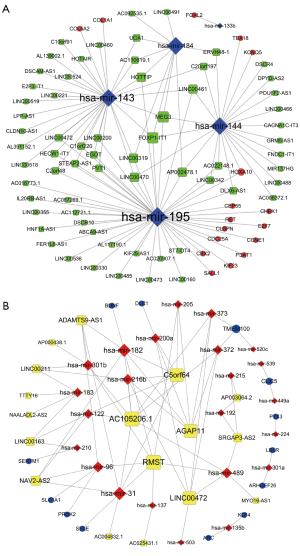
On the basis of the above data, lncRNA-miRNA-mRNA ceRNA network was constructed and visualized using Cytoscape software. On the basis of ceRNA hypothesis and the expression levels of DElncRNAs, DEmiRNAs and DEmRNAs, two lncRNA-miRNA-mRNA ceRNA networks including over-expressed (Figure 2A) and under-expressed (Figure 2B) networks were constructed and visualized using Cytoscape. In over-expressed ceRNA network, 65 up-regulated DElncRNAs, 5 down-regulated DEmiRNAs and 17 up-regulated DEmRNAs were included and 114 edges were contained. In under-expressed ceRNA network, 17 down-regulated DElncRNAs, 24 up-regulated DEmiRNAs and 13 down-regulated DEmRNAs were included and 78 edges were contained.
Construction of PPI network and key gene validation
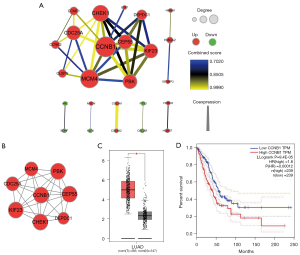
The interactive relationships of 57 DEmRNAs encoding proteins in ceRNA network were elucidated using PPI network. At minimum required interaction score = the high confidence 0.7, a total of 25 among 57 genes was filtered into PPI network, and a PPI network with 25 nodes and 38 edges was established (Figure 3A). Highly correlated module analysis showed that 2 modules were identified in whole PPI network, and the most significant module (subnetwok1) included 8 nodes and 26 edges (Figure 3B). Centrality analysis showed that centrality score of CCNB1 gene obtained from each centrality method ranked first (degree =7, betweenness centrality =0.01904762, closeness centrality =1, eigenvector =0.407759339, subgraph =73.9836731, information =2.274614334, network =6.647619048), and CCNB1 gene was identified as essential gene in subnetwork1. The expression analysis based on GEPIA database showed CCNB1 gene was significantly up-regulated in LUAD (Figure 3C), and survival analysis revealed that low mRNA expression of CCNB1 resulted in a higher OS rate than high mRNA expression in LUAD patients (Figure 3D). CCNB1 is the Protein Coding gene of Cyclin B1, and play role in mitosis. Cyclin B1 is necessary for proper control of the G2/M transition phase of the cell cycle. Some studies have reported that the dysregulated expression of CCNB1 resulted in many diseases including cancer (39,40). Recently, several studies showed that overexpression of CCNB1 was correlated with poor survival in most solid tumors including LUAD (41,42), which suggested that the expression status of CCNB1 was able to serve as a significant prognostic predictor in solid tumors (42). Our result further validated the feasibility of mRNA CCNB1 as a prognosticator in LUAD.
Gene function analysis of DEmRNAs
For the purpose of better understanding the roles of the DElncRNAs in ceRNA network in LUAD, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for 57 DEmRNAs in ceRNA network were performed using online STRING database, and GO terms and KEGG pathways regulated by DElncRNAs were identified. Biological process (BP), cellular component (CC) and molecular function (MF) of the most significant association to DEmRNAs were separately positive regulation of biological process [GO: 0048518, false discovery rate (FDR) =5.12E-05], chromatin (GO: 0000785, FDR =0.0137) and sequence-specific DNA binding (GO: 0043565, FDR =0.00101). The most significant enrichment KEGG pathway was cell cycle (has04110, FDR =0.000308). As we all know, cell cycle is one of the most important pathways belonging to cell growth and death, and lots of studies have demonstrated that cell cycle was dysregulated in various cancers (43,44).
OS analysis of LUAD-specific lncRNA signature
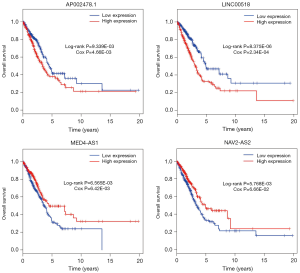
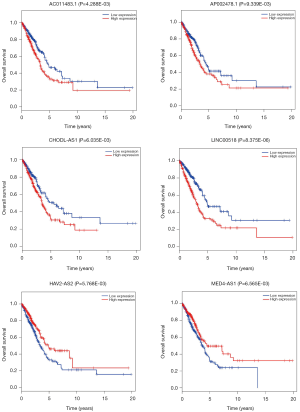
The relationships of OS of LUAD patients and DElncRNAs in ceRNA network were determined using KM and LR test. According to P<0.01 cut-off threshold, 9 DElncRNAs including LINC00518 (P=8.375E-06), UCA1 (P=4.709E-03), NAV2-AS2 (P=5.768E-03), CHODL-AS1 (P=6.035E-03), MED4-AS1 (P=6.565E-03), SYNPR-AS1 (P=9.765E-04), AC011483.1 (P=4.288E-03), POU6F2-AS1 (P=7.019E-03) and AP002478.1 (P=9.339E-03) were found to be related to OS of LUAD-patients (Figure S1).
Establishment of lncRNA signature prognostic model
We used univariate regression analysis to identify DElncRNAs associated with the OS of LUAD patients. With P<0.01 cut-off threshold, a group of 18 DElncRNAs including LINC00518 (P=5.11E-06), C20orf197 (P=8.68E-05), AP002478.1 (P=0.000365572), C11orf44 (P=0.000933011), LINC00460 (P=0.001760147), UCA1 (P=0.001836663), SYNPR-AS1 (P=0.002361183), LINC00319 (P=0.003555803), LINC00337 (P=0.00423919), AC004832.1 (P=0.004503163), LNX1-AS2 (P=0.004812281), NAV2-AS2 (P=0.004889676), C5orf64 (P=0.00513461), AC080129.1 (P=0.006289307), MED4-AS1 (P=0.006517782), AP000438.1 (P=0.008252988), AC011483.1 (P=0.008263272) and ABCA9-AS1 (P=0.009646077) was detected to have significantly prognostic value (http://fp.amegroups.cn/cms/tcr.2019.06.09-4.pdf). Further, we found that 7 DElncRNAs (LINC00518, UCA1, NAV2-AS2, MED4-AS1, SYNPR-AS1, AC011483.1, AP002478.1) were simultaneously identified to be related to OS in KM (LR test) and univariate Cox regression analysis. The 7 DElncRNAs were fitted into the multivariate Cox regression model, and the result showed that a group of 4 DElncRNAs including AP002478.1 (Cox P=4.66E-03), LINC00518 (Cox P=2.34E-04), MED4-AS1 (Cox P=6.42E-03) and NAV2-AS2 (Cox P=6.66E-02) had significantly prognostic value with OS of LUAD patients (Figure 4), and 4-lncRNA prognostic model was established. With the risk score of each patient (http://fp.amegroups.cn/cms/tcr.2019.06.09-5.pdf), the patients were divided into high- and low- risk groups (Figure 5A,B). Comparing to low risk group, the mortality rate of patients in high-risk group was significantly higher, and high-risk group was related with worse prognosis (Figure 5C). The ROC of 3-year survival correlation of the 4-lncRNA signature was analyzed, and area under the curve (AUC) was computed. The AUC of 4-lncRNA signature was 0.669, which indicated its effectiveness as a prognostic biomarker in evaluating OS of LUAD-patients (Figure 5D).
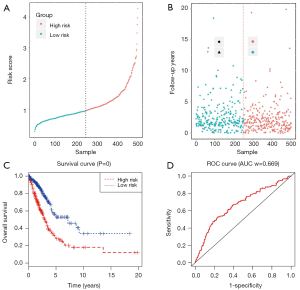
The expressions of the 4 lncRNAs were analyzed in LUAD tissues and normal lung tissues (Figure 6A), in high-risk and low-risk patient groups (Figure 6B), and in alive and dead groups (Figure 6C). AP002478.1 and LINC00518 were expressed at high level in LUAD patients with low risk scores. Whereas, MED4-AS1 and NAV2-AS2 were expressed at low level in LUAD patients with high risk scores. The expressions of 4 lncRNAs in alive and dead patients showed that AP002478.1 and LINC00518 were expressed at high level, and MED4-AS1 and NAV2-AS2 were expressed at low level in dead patients. The expressions [log2 (expression value +1)] of the 4 lncRNAs were showed in Figure 6D.
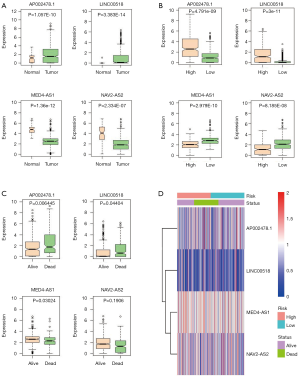
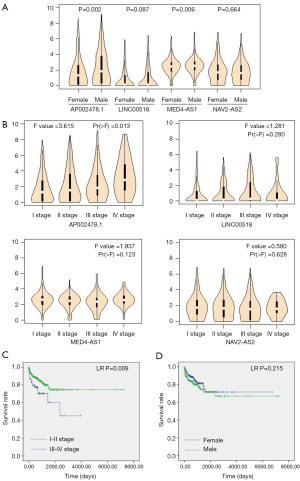
Correlation of four-lncRNA signature and clinical factor
The expression analysis of 4-lncRNA signature showed that AP002478.1 and MED-AS1 were highly expressed in male LUAD patients (P=0.002, 0.006, respectively; Figure 7A), and expression of AP002478.1 was significantly correlated to pathological stage and was increased with pathological stage (P=0.013; Figure 7B). Kaplan-Meier analysis showed that pathological stage was significantly correlated to OS of LUAD patients (LR P=0.009; Figure 7C), and the correlation of gender and OS had no statistical significance (LR P=0.215; Figure 7D). The results indicated that pathological stage was independent prognostic factor of OS in LUAD patients.
Discussion
LC is among the most common malignant cancer, and has jumped to the first cause of cancer-related malignant in recent years. LUAD is the major pathological subtype of LC (7), but the majority of the patients with LUAD remain short survival times, and the overall 5-year survival rate for all stages of LUAD less than 20% (4). The identifying specific diagnostic and prognostic biomarkers may contribute to improve the OS rate of LUAD patients. For this, it is necessary to further understand the regulatory mechanisms of LUAD initiation and progression, and to identify potentially prognostic signatures related to LUAD. Increasing evidence shows that lncRNAs acting as ceRNAs play key roles in cancer biology (10).
In recent years, several studies have explored the roles of RNAs in LUAD-specific ceRNA network (20,45,46). For example, Li et al. investigated the LUAD-related ceRNA network and identified some potential LUAD-related prognostic lncRNAs using univariate Cox regression (46), but did not consider multivariate Cox regression. Similarly, another study also analyzed the LUAD-related ceRNA network and validated some lncRNAs (45), but still did not take into account the prognostic signature using multivariate Cox regression. In this study, we integrated LUAD-related lncRNA, miRNA and mRNA expression data from TCGA database and constructed LUAD-specific lncRNA-miRNA-mRNA ceRNA network based on the ceRNA theory. Further, we investigated the association of lncRNAs in ceRNA network and OS of LUAD patients. KM (LR test) and univariate regression analysis showed that 7 lncRNAs in ceRNA network were simultaneously associated with OS of LUAD patients. Multivariate regression analysis showed that prognostic value of 4 lncRNAs (AP002478.1, LINC00518, MED4-AS1 and NAV2-AS2) in those 7 lncRNAs had statistical significance in the OS of LUAD patients. A cumulative risk score of the 4 lncRNAs was calculated, and 0.669 AUC indicated that the 4-lncRNA signature might predict and evaluate the OS of LUAD patients.
In the current study, lncRNAs AP002478.1, LINC00518, MED4-AS1 and NAV2-AS2 were identified to play important roles in the prognosis of LUAD patients. A recent study showed that the expression of AP002478.1 was associated with GC with Helicobacter pylori infection, and might serve as a prognostic biomarker of OS in GC patients with positive Helicobacter pylori (47). In this study, we noticed that the expression of AP002478.1 was significantly up-regulated in LUAD tissues, and up-regulated AP002478.1 could compete with down-regulated miRNAs (has-mir-195, has-mir-144, has-mir-184) to regulate the expression of the target genes such as CHEK1, KIF23, CCNE1 and HOXA10 involved in over-expressed ceRNA network (Figure 2A). Previous studies have reported that those miRNAs and their targets were associated with cancers (48-53). For example, Zhao et al. found that down-regulated has-mir-195 was associated with poor differentiation of NSCLC (50), and Tan et al. found down-regulated has-mir-195 was related to the onset, erosion and transfer of colorectal cancer (54). A lot of studies reported that has-mir-144 were aberrantly expressed in some cancers such as osteosarcoma (55), rectal cancer (56), and cervical cancer (57). In addition, several studies showed has-mir-184 was down-regulated in epithelial ovarian cancer and could act as a potentially diagnostic and prognostic marker (58). So far, no study has reported that LINC00518, MED4-AS1 and NAV2-AS2 were associated with cancers. The present study is the first to identify the aberrant expression of AP002478.1, LINC00518, MED4-AS1 and NAV2-AS2 in LUAD, and indicated the potential roles of the 4-lncRNA signature as prognostic biomarker in LUAD. This study will contribute to further understand the lncRNA-mediated ceRNA regulatory mechanisms in LUAD, and identify novel lncRNAs as potentially prognostic biomarkers and/or therapeutic targets.
Although the findings of this study provided novel potential important lncRNA biomarkers for clinic in predicting and evaluating OS of LUAD patients, the limitations were that the results were not verified by other experimental methods. Further, the biological roles of the 4 lncRNAs in LUAD need to be investigated.
Conclusions
In the current study, 4-lncRNA signature was identified as a potentially prognostic predictor of OS for LUAD patients based on ceRNA network. The current findings provide some novel insights into the regulatory mechanisms related to lncRNA-related ceRNA network in LUAD and identify 4 lncRNAs to serve as potentially prognostic biomarkers. Further, functional studies underlying lncRNAs function in LUAD-specific ceRNA network are required to elucidate.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All RNAseq data and clinical information were obtained from public TCGA database, and were approved by the Institutional Review Board of the relevant participating institutions and by the National Cancer Institute of NIH. No approval from the ethics committee was required. The present study meets the requirements of data usage and publishing from TCGA database. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Yao T, Sung HY, Mao Z, et al. The healthcare costs of secondhand smoke exposure in rural China. Tob Control 2015;24:e221-6. [Crossref] [PubMed]
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017; [Crossref] [PubMed]
- Wu K, House L, Liu W, et al. Personalized targeted therapy for lung cancer. Int J Mol Sci 2012;13:11471-96. [Crossref] [PubMed]
- Walters S, Maringe C, Coleman MP, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax 2013;68:551-64. [Crossref] [PubMed]
- Nesbitt JC, Putnam JB Jr, Walsh GL, et al. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg 1995;60:466-72. [Crossref] [PubMed]
- Zagryazhskaya A, Gyuraszova K, Zhivotovsky B. Cell death in cancer therapy of lung adenocarcinoma. Int J Dev Biol 2015;59:119-29. [Crossref] [PubMed]
- Ohba T, Toyokawa G, Osoegawa A, et al. Mutations of the EGFR, K-ras, EML4-ALK, and BRAF genes in resected pathological stage I lung adenocarcinoma. Surg Today 2016;46:1091-8. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013;34:613-20. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet 2010;19:R152-61. [Crossref] [PubMed]
- Hao F, Mou Y, Zhang L, et al. LncRNA AFAP1-AS1 is a prognostic biomarker and serves as oncogenic role in retinoblastoma. Biosci Rep 2018; [Crossref] [PubMed]
- Qin J, Ning H, Zhou Y, et al. LncRNA MIR31HG overexpression serves as poor prognostic biomarker and promotes cells proliferation in lung adenocarcinoma. Biomed Pharmacother 2018;99:363-8. [Crossref] [PubMed]
- Xiao Z, Qu Z, Chen Z, et al. LncRNA HOTAIR is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ss-Catenin Signaling Pathway. Cell Physiol Biochem 2018;46:1275-85. [Crossref] [PubMed]
- Zeng Y, Wang T, Liu Y, et al. LncRNA PVT1 as an effective biomarker for cancer diagnosis and detection based on transcriptome data and meta-analysis. Oncotarget 2017;8:75455-66. [Crossref] [PubMed]
- Liu C, Ren L, Deng J, et al. LncRNA TP73-AS1 promoted the progression of lung adenocarcinoma via PI3K/AKT pathway. Biosci Rep 2019;39. [PubMed]
- Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 Accelerates Lung Adenocarcinoma Deterioration and Binds to Mir-193a-3p as a Competitive Endogenous RNA. Cell Physiol Biochem 2018;48:905-18. [Crossref] [PubMed]
- Ye JJ, Cheng YL, Deng JJ, et al. LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma by targeting miR-302c-5p/FOXA1 axis. Gene 2019;685:76-84. [Crossref] [PubMed]
- Li L, Feng T, Qu J, et al. LncRNA Expression Signature in Prediction of the Prognosis of Lung Adenocarcinoma. Genet Test Mol Biomarkers 2018;22:20-8. [Crossref] [PubMed]
- Li X, Li B, Ran P, et al. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett 2018;16:5697-708. [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Conte F, Fiscon G, Chiara M, et al. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One 2017;12:e0171661. [Crossref] [PubMed]
- Liu H, Zhang Z, Wu N, et al. Integrative Analysis of Dysregulated lncRNA-Associated ceRNA Network Reveals Functional lncRNAs in Gastric Cancer. Genes (Basel) 2018;9:303. [Crossref] [PubMed]
- Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer 2018;17:119. [Crossref] [PubMed]
- Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med 2018;16:264. [Crossref] [PubMed]
- Zhang Z, Wang S, Ji D, et al. Construction of a ceRNA network reveals potential lncRNA biomarkers in rectal adenocarcinoma. Oncol Rep 2018;39:2101-13. [PubMed]
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 2013;3:1113-21. [Crossref] [PubMed]
- Jin X, Guan Y, Sheng H, et al. Crosstalk in competing endogenous RNA network reveals the complex molecular mechanism underlying lung cancer. Oncotarget 2017;8:91270-80. [Crossref] [PubMed]
- Wei Y, Chang Z, Wu C, et al. Identification of potential cancer-related pseudogenes in lung adenocarcinoma based on ceRNA hypothesis. Oncotarget 2017;8:59036-47. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Guo LL, Song CH, Wang P, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol 2015;21:11680-7. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Zeng JH, Liang L, He RQ, et al. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget 2017;8:16811-28. [PubMed]
- Chai N, Xie HH, Yin JP, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun 2018;500:924-9. [Crossref] [PubMed]
- Liu D, Xu W, Ding X, et al. Polymorphisms of CCNB1 Associated With the Clinical Outcomes of Platinum-Based Chemotherapy in Chinese NSCLC Patients. J Cancer 2017;8:3785-94. [Crossref] [PubMed]
- Ding K, Li W, Zou Z, et al. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses 2014;83:359-64. [Crossref] [PubMed]
- Ye C, Wang J, Wu P, et al. Prognostic role of cyclin B1 in solid tumors: a meta-analysis. Oncotarget 2017;8:2224-32. [PubMed]
- Fan Y, Zhu X, Xu Y, et al. Cell-Cycle and DNA-Damage Response Pathway Is Involved in Leptomeningeal Metastasis of Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:209-16. [Crossref] [PubMed]
- Sherr CJ. Cell cycle control and cancer. Harvey Lect 2000-2001;96:73-92. [PubMed]
- Sui J, Li YH, Zhang YQ, et al. Integrated analysis of long non-coding RNAassociated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol 2016;49:2023-36. [Crossref] [PubMed]
- Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci 2016;20:2285-95. [PubMed]
- Liu Y, Zhu J, Ma X, et al. ceRNA network construction and comparison of gastric cancer with or without Helicobacter pylori infection. J Cell Physiol 2019;234:7128-40. [Crossref] [PubMed]
- Wang YB, Zhao XH, Li G, et al. MicroRNA-184 inhibits proliferation and promotes apoptosis of human colon cancer SW480 and HCT116 cells by downregulating C-MYC and BCL-2. J Cell Biochem 2018;119:1702-15. [Crossref] [PubMed]
- Yao Q, Gu A, Wang Z, et al. MicroRNA-144 functions as a tumor suppressor in gastric cancer by targeting cyclooxygenase-2. Exp Ther Med 2018;15:3088-95. [PubMed]
- Zhao Q, Cao J, Wu YC, et al. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res 2015;5:1692-705. [PubMed]
- Kato T, Wada H, Patel P, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer 2016;92:53-61. [Crossref] [PubMed]
- Noack S, Raab M, Matthess Y, et al. Synthetic lethality in CCNE1-amplified high grade serous ovarian cancer through combined inhibition of Polo-like kinase 1 and microtubule dynamics. Oncotarget 2018;9:25842-59. [Crossref] [PubMed]
- Yuan Y, Sun S, Jiao N, et al. Upregulation of HOXA10 Protein Expression Predicts Poor Prognosis for Colorectal Cancer. Genet Test Mol Biomarkers 2018;22:390-7. [Crossref] [PubMed]
- Tan YG, Zhang YF, Guo CJ, et al. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med 2013;6:972-6. [Crossref] [PubMed]
- Green D, Mohorianu I, Piec I, et al. MicroRNA expression in a phosphaturic mesenchymal tumour. Bone Rep 2017;7:63-9. [Crossref] [PubMed]
- Cai SD, Chen JS, Xi ZW, et al. MicroRNA144 inhibits migration and proliferation in rectal cancer by downregulating ROCK1. Mol Med Rep 2015;12:7396-402. [Crossref] [PubMed]
- Tao P, Wen H, Yang B, et al. miR-144 inhibits growth and metastasis of cervical cancer cells by targeting VEGFA and VEGFC. Exp Ther Med 2018;15:562-8. [PubMed]
- Qin CZ, Lou XY, Lv QL, et al. MicroRNA-184 acts as a potential diagnostic and prognostic marker in epithelial ovarian cancer and regulates cell proliferation, apoptosis and inflammation. Pharmazie 2015;70:668-73. [PubMed]


