
Downregulated let-7d positively stimulates rectum adenocarcinoma cell malignant biological behaviors by upregulating ATP binding cassette subfamily C member 2
Introduction
Rectum adenocarcinoma is one of the most common malignant tumors in the digestive tract (1). In present, the pathogeny of rectum adenocarcinoma is still enigmatic, its occurrence mainly related to social environment, dietary habits and genetic factors (2). Up to now, substantial progresses have been made in diagnosis and treatment of rectum adenocarcinoma (3). However, because of its deep position in the pelvic cavity, the surgery is not easy to be complete, resulting in a high postoperative recurrence rate (4). In recent years, molecular therapies directing at rectum adenocarcinoma have attracted increasing attentions, while to date none molecular biomarkers have yet reached the clinic (5). Therefore, it is urgent to explore accurate targeted molecules for diagnosis and prognosis of rectum adenocarcinoma.
MicroRNA (miRNA) is a kind of naturally generated and highly conserved small non-coding RNA with a length of about 22 nucleotides, which can interact with 3' untranslation region (UTR) of target mRNA according to sequence complementarity, leading to degradation or translation inhibition of mRNA (6). Meanwhile, miRNA regulates proliferation, invasion and apoptosis of cancer cells in different up or down regulated ways, and also affects the expression of genes involved in signal transduction of cancer cells (7). Let-7d, widely considered to be a tumor suppressor of miRNA, has been found to be highly conserved in many species, including humans (8). Childs et al. found that let-7d was low expressed in head and neck squamous cell carcinoma (HNSCC) patients, and easily led to poor survival(9). Shell et al. demonstrated that loss of let-7d was more likely to result in unfavorable prognosis with the patients in ovarian cancer (10). Similarly, down-regulation of let-7d was also found in other cancers including prostate cancer (11), breast cancer (12), colorectal cancer (13), etc. However, the function of let-7d in rectum adenocarcinoma has not been described.
In this research, we explored that let-7d was down-regulated both in rectum adenocarcinoma tissues and cells. The vitro experiment proved that over-expression of let-7d could suppress the proliferation, invasion and migration of SW837 cells. Then, the direct interaction between let-7d and ATP binding cassette subfamily C member 2 (ABCC2) was confirmed by the dual luciferase assay. Moreover, we found that ABCC2 was highly expressed in patients with rectum adenocarcinoma, and its high expression easily led to worse overall survival. Co-transfection assay testified that silencing ABCC2 suppressed the proliferation, invasion and migration of SW837 cells, meanwhile accentuated the inhibitory effect of let-7d on SW837 cells. Herein, all results of this study showed that let-7d regulated ABCC2 as a crucial modulator in the progression of rectum adenocarcinoma, which may provide a new insight for the treatment of rectum adenocarcinoma.
Methods
Data collection
The differential expression analysis of let-7d (including 162 human tumor tissues and 3 normal tissues) and ABCC2 (including 167 human tumor tissues and 10 normal tissues) were got from TCGA database (https://cancergenome.nih.gov/). And then the overall survival rate was plotted and Cox regression analyze was performed.
Cell culture and transfection
Rectum adenocarcinoma cell lines SW837 and normal cell line Hs680.Rec were purchased from the Shanghai cell bank of the Chinese academy of medical sciences. All the cells were incubated in Dulbecco’s Eagle medium with 10% fetal bovine serum (FBS), under the condition of 37 °C, 5% CO2. When cells came into the logarithmic phase, the let-7d mimic/mimic NC (RIBOBIO, Guangzhou, China), si-ABCC2 and si-con (Gene-Pharma, Shanghai, China) were transfected into the cells using the Lipofectamine 2000 transfection kit according to the supplier’s protocol. The siRNAs sequences were presented as follows: si-ABCC2: 5'-GCACCATCTTAGAGAAGGGAT-3', si-con: 5'-AATTCTCCGAACGTGTCACGT-3'.
Reverse transcription quantitative polymerase chain reaction (qRT-PCR)
To get the relative RNA expression of let-7d and ABCC2, total RNA was isolated from the transfected cells by TRIzol solution (Invitrogen, Carlsbad, CA, USA) and then qRT-PCR has been taken. Subsequently, the RNA was reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen; Carlsbad, CA, USA). qRT-PCR was performed using SsoFast™ EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s procedure and in a real-time thermal cycler: 40 cycles including 95 °C for 5 min, 95 °C for 30 s, and an extension step at 60 °C for 45 s and 72 °C for 30 min; 2−ΔΔCT method was used to measure the relative expression of let-7d and ABCC2, GAPDH was as the internal control and the primer sequences were as follows: let-7(D) F: 5'-UUGGUGUGUUGGAUGAUGGAGU-3', R: 5'-AACGCTTCACGAATTTGCGT-3'. ABCC2: F: 5'-GTTCCAAGGAGAAGCCCACA-3', R: 5'-CCGGTCCACATTCTCTGGAA-3'. GAPDH: F: 5'-GGAGCGAGATCCCTCCAAAAT-3', R: 5’-GGCTGTTGTCATACTTCTCATGG-3'.
Western blot
Total protein was extracted from the transfected cells using RIPA regent (Beyotime, Shanghai, China) with 1% protease inhibitor, quantified through BCA Protein Assay Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), separated by 12% SDS-PAGE and electrotransferred onto PVDF membranes. Subsequently, the membranes were blocked with 5% skimmed milk powder for 1 h and against with primary antibodies (Abcam, Cambridge, MA, USA), ABCC2 (ab203397, 1:1,000), GAPDH (ab9485, 1:1,000) overnight while incubated with secondary antibody (Abcam, Cambridge, MA, USA, ab150077, 1:2,000) for 1 h. Finally, added ECL reagent (Beyotime, Jiangsu, China) to detect the signals, and scanned with QUANTITY ONE.
CCK-8 assay
The transfected cells were prepared for cell suspension, seeded into 96-well plates at the standard of 1,000 cells per well and cultured in a carbon dioxide incubator. Cell activity was detected at 0 h, 24 h, 48 h and 72 h, added 10 µL of CCK-8 reagent into each well and cultured in 37 °C incubator for 1.5 h before each detection. Using microplate reader (Bio-Rad, Hercules, CA, USA) to evaluate the optical density (OD) value and draw the proliferation curve.
Transwell assay
Transwell assay was used to measure the cells migration and invasion abilities. Put the transfected cells into suspension and added it into the upper chamber. Meanwhile, 500 µL of complete medium was filled into the under chamber. Overnight, swabbed remaining cells in upper chamber, washed by PBS, fixed with 4% paraformaldehyde for 30 min, dyed through 0.1% crystal violet for 20 min and captured via microscope. It is worth noting that the upper chamber of the invasive detection needs to be pre-coated with Matrigel (BD Sciences, Franklin Lakes, NJ, USA) while migratory experiment was not. Furthermore, the invasion (1×105) density of inoculated cells was different from migration (5×103).
Luciferase reporter assay
Dual luciferase report was further confirmed the prediction results from the bioinformatics software, and dual luciferase reporting kit was used with Dual Luciferase™ reporting system. A reporter plasmid wild-type ABCC2 (WT ABCC2) was conducted by inserting pmirGLO Vector (Promega, USA) with ABCC2 sequences, which includes the target site of let-7d. The target site was mutated and inserted to form a negative control for reporter plasmid, which was called mutated-type ABCC2 (MUT ABCC2). The let-7d mimic/NC and vector were co-transfected into the cells through Lipofectamine 2000 transfection kit according to the supplier’s instruction. After 48 h, the proteins in the cells were extracted for luciferase detection. In the end, using a Dual Luciferase™ reporting system to detect the firefly luciferin to Marine luciferin intensity, assess the relative luciferase activity and reflect the effect of let-7d on ABCC2 level.
Statistical analysis
To analyze the experiment data using SPSS 22.0 and GraphPad Prism 5.0 Software. The comparison between the two groups was used Student’s t-test and ANOVA analysis followed by Dunnet’s (compare all groups vs. control group) and Bonferroni’s post hoc test for multiple groups (compare selected pairs of groups). The overall survival was analyzed through Kaplan-Meier method and log-rank test. The independent prognostic analysis was performed using Cox regression. P<0.05 was considered statistically significant.
Results
Let-7d is down-regulated in rectum adenocarcinoma
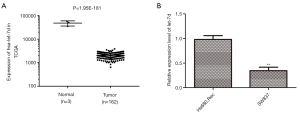
The data from the TCGA database showed that the expression of let-7d was significantly decreased in tumor tissues compared with the normal (Figure 1A, P=1.95E-181). To further explore the function of let-7d, we examined its expression in rectum adenocarcinoma cell line SW837. The results indicated that let-7d was down-regulated in rectum adenocarcinoma cell line SW837 in contrast with normal cell line Hs680.Rec (Figure 1B, **P<0.01). So, SW837 cells were used for the subsequent experiments. The lower expression of let-7d in rectum adenocarcinoma samples insinuated that let-7d served on an important role in the progression of rectum adenocarcinoma.
Let-7d suppresses the proliferation and mobility of rectum adenocarcinoma cells
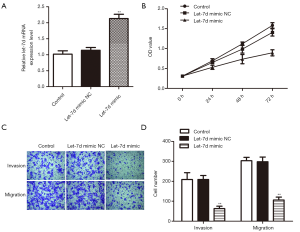
To investigate the effects of let-7d on rectum adenocarcinoma, let-7d mimic was used to up-regulate let-7d in rectum adenocarcinoma cell. Results of qRT-PCR analysis showed that the relative expression of let-7d was remarkably improved after transfected with let-7d mimic (Figure 2A, **P<0.01) compared with the control and let-7d mimic NC groups. Then, CCK-8 assay demonstrated that high expression of let-7d significantly reduced the rectum adenocarcinoma cells OD values (Figure 2B, **P<0.01) and transwell assay exhibited that overexpression of let-7d obviously decreased the migrated and invaded cells number in contrast with let-7d mimic NC and control groups (Figure 2C,D, **P<0.01). The findings manifested that up-regulation of let-7d could repress rectum adenocarcinoma cells proliferative, invasive and migratory capacities.
ABCC2 is an immediate target of let-7d
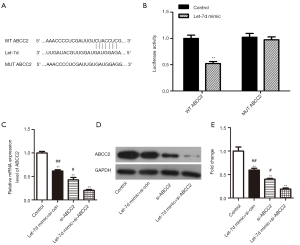
According to research reports, miRNAs performed their function via restraining the expression of their target genes. Therefore, in order to further explore let-7d molecular mechanism, miRNA target analysis tools TargetScan, MiRanda, miRDB and miRWalk were used to predict the possible binding site to ABCC2. As bioinformatics results shown, a diffusely-conserved binding site between 3' UTR of ABCC2 and let-7d was be found and confirmed by luciferase reporter assay in HEK293T cells, meanwhile the mutant ABCC2 sequence was cloned into luciferase reporter vectors (Figure 3A). Subsequently, let-7d mimic and control were transfected into HEK293T cells, compared with the control, we found that the luciferase reporter activity was obviously declined with high-expression of let-7d in WT ABCC2 group. But the luciferase reporter activity was almost unchanged in MUT ABCC2 group (Figure 3B, **P<0.01). Finally, let-7d mimic and si-ABCC2 were transfected into SW837 cells to explore the effect on ABCC2. As the Figure 3C,D,E shown, qRT-PCR and western blot indicated that ABCC2 expression was significantly down-regulated after transfected with let-7d mimic + si-con, si-ABCC2, or let-7d mimic + si-ABCC2 compared with control, especially down-regulated in let-7d mimic + si-ABCC2 group (**P<0.01, #P<0.05, ##P<0.01). Taken together, the findings provided an evident testimony indicating that ABCC2 was a special target of let-7d and negatively regulated by let-7d.
High expression of ABCC2 easily leads to worse overall survival in rectum adenocarcinoma
To further explore the function of ABCC2 in rectum adenocarcinoma, a comprehensive study was performed. The data from TCGA database and Kaplan Meier-plotter websites showed that ABCC2 was significantly over-expressed in tumor tissues (Figure 4A, P=1.11E-0.5) and overexpression of ABCC2 was more easily led to poor overall survival with the patients in rectum adenocarcinoma (Figure 4B, P=0.03). Moreover, as shown in Table 1, the Cox regression analysis showed that the prognostic significance of rectum adenocarcinoma patients was correlated with several parameters, in addition to ABCC2 expression. Univariate analysis found that ABCC2 expression (high/low), clinical-stage, pathologic-M/N and age (<60/≥60) were involved in the prognosis of rectum adenocarcinoma. Then, these elements were studied again in multivariate analysis, and indicated that only age (<60/≥60) can be used as an independent prognostic indicator of patient with rectum adenocarcinoma. The consequence indicated that up-regulation of ABCC2 was observed in rectum adenocarcinoma patients and its elevation resulted in worse prognosis.
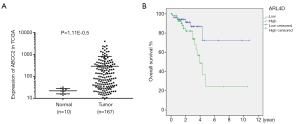
Table 1
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| P | HR | 95%CI | P | HR | 95%CI | ||
| ABCC2 expression (high/low) | 0.046* | 2.488 | 1.017–5.892 | 0.364 | 1.986 | 0.599–4.033 | |
| Clinical-Stage (I + II/III + IV) | 0.009* | 3.517 | 1.364–9.072 | 0.947 | 0.000 | 1.770E+96 | |
| Pathologic-T (T1 + T2/T3 + T4) | 0.465 | 1.498 | 0.507–4.426 | – | – | – | |
| Pathologic-M (M0/M1) | 0.007* | 3.342 | 1.395–8.008 | 0.453 | 2.238 | 0.530–4.145 | |
| Pathologic-N (N0/N1 + N2 + N3) | 0.011* | 3.149 | 1.296–7.650 | 0.939 | 7247.337 | 0.000–3.222E+103 | |
| Age (<60/≥60 years) | 0.013* | 6.300 | 1.476–26.899 | 0.030* | 9.394 | 1.243–71.003 | |
| Gender (female/male) | 0.811 | 0.908 | 0.413–2.000 | – | – | – | |
*, P<0.05. HR, hazard ratio; CI, confidence interval; T, tumor; N, lymph nodes; M, metastasis; ABCC2, ATP binding cassette subfamily C member 2.
Deleted expression of ABCC2 enhanced the inhibitory effect of let-7d on SW837 cells
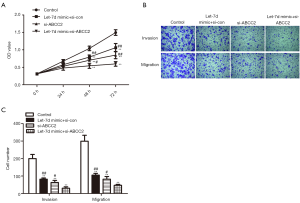
Based on the above research, we speculated that the repressive role of let-7d overexpression on proliferation, invasion and migration of rectum adenocarcinoma cells was correlated with ABCC2 involvement. Initially, let-7d mimic and si-ABCC2 were transfected into SW837 cells to detect the effect of let-7d/ABCC2 on rectum adenocarcinoma development. The data from CCK-8 assay showed that the OD value was obviously declined in let-7d mimic + si-con, si-ABCC2, and let-7d mimic + si-ABCC2 groups compared with the control, especially declined in let-7d mimic + si-ABCC2 group, indicating that down-regulation of ABCC2 can accelerate the suppression effect of let-7d on the proliferation of SW837 cells (Figure 5A, #P<0.05, ##P<0.01, **P<0.01). Transwell assay discovered that the number of invaded and migrated cells were significantly reduced in let-7d mimic + si-con, si-ABCC2, and let-7d mimic + si-ABCC2 groups compared with the control, especially reduced in let-7d mimic + si-ABCC2 group, suggesting that loss ABCC2 increased the inhibitory effect of let-7d on SW837 cells invasion and migration (Figure 5B,C, #P<0.05, ##P<0.01, **P<0.01). In conclusion, the data indicated that down-regulation of ABCC2 increased the restraining of SW837 cells proliferation, invasion and migration caused by let-7d, demonstrated that let-7d acted as a potential suppressor in rectum adenocarcinoma by targeting ABCC2 (**P represented vs. control group, #P and ##P represented vs. let-7d mimic + si-ABCC2 group).
Discussion
Our data showed that let-7d was down-regulated in rectum adenocarcinoma samples, CCK-8 and transwell assays indicated that over-expression of let-7d positively suppressed SW837 cells proliferation, invasion and migration. We also testified that ABCC2 was a direct functional target of let-7d through prediction software and dual luciferase reporter assay. The analysis from TCGA investigated that ABCC2 was highly expressed in rectum adenocarcinoma tissues and its high expression level tended to poor overall survival. Co-transfection assay found that knockdown ABCC2 could emphasize the inhibitory effect of let-7d on SW837 cells. Taken together, the study indicated that over-expression of let-7d repressed rectum adenocarcinoma progression through targeting with ABCC2.
Emerging evidence pointed that let-7d was down-regulated in numerous cancers and acted as a tumor-inhibiting factor (8). For instance, let-7d was found to be reduced in oral squamous cell carcinoma (14), ovarian cancer (15) and down-regulation of let-7d could suppress proliferation, invasion and migration of renal cell carcinoma (16). These findings were consistent with our research in rectum adenocarcinoma. In our study, we found that let-7d was obviously reduced in rectum adenocarcinoma samples and up-regulation of let-7d could repress SW837 cells proliferation, invasion and migration. Let-7d has been reported to be involved in the pathogenesis of some cancers (17), for example, let-7d was found to have anti-tumor and carcinogenic effects in osteosarcoma (18). In addition, let-7d can also directly target CCL7, COL3A1 (19), HMGA2 and PBX3, and suppress cell proliferation, invasion and metastasis. These evidence not only provide strong supports for the influence of let-7d in rectum adenocarcinoma in our research, but also provide new strategy for the treatment of rectum adenocarcinoma.
The modulation of let-7d by downstream target genes acted as a crucial role in exhibiting its anti-cancer mechanism. Therefore, multiple bioinformatics prediction software was used to analyze the downstream gene of let-7d and confirmed it with dual luciferase reporter assay. The results showed that ABCC2 was a vital downstream target gene of let-7d. ABCC2, as a member of the ATP binding cassette superfamily, also known as multidrug resistance-associated protein 2 (MRP2), encodes human canalicular multi-specific organic anion transporter (20). In addition to being expressed in normal human tissues, ABCC2 was also detected in numerous tumors, including hepatocellular (21), ovarian (22), lung (23), breast (24), gastric (25) and colorectal carcinomas (26). For instance, ABCC2 is also shown to increase cisplatin resistance through targeting let-7c in non-small cell lung cancer (27) and low expression of ABCC2 was related to longer overall survival in ovarian carcinoma (28). While, the function of this gene in rectum adenocarcinoma has been poorly studied. The results exhibited that ABCC2 was over expressed in rectum adenocarcinoma tissues and led to worse prognosis. Meanwhile, the study also found that ABCC2, as the downstream target gene, interacted with let-7d to suppress the rectum adenocarcinoma cells growth and movement.
In conclusion, to the best of our knowledge, this is the first time to discover that let-7d repressed rectum adenocarcinoma development through targeting ABCC2. The study indicated that let-7d was lower expression in rectum adenocarcinoma tissues and cells, and in vitro experiments showed that up-regulation of let-7d could significantly inhibit SW837 cells proliferation, invasion and migration. While, ABCC2 was found highly expressed in rectum adenocarcinoma tissues and easily led to poor prognosis. The potential anti-cancer function of let-7d was mediated by declining the ABCC2 as its target gene. However, every miRNA may regulate many target genes in different ways, affecting the occurrence and progression of cancer, thus more detailed studies on the molecular and cellular mechanism of let-7d target were required.
Conclusions
This study provides a novel insight for the treatment of rectum adenocarcinoma.
Acknowledgments
Funding: This study was supported by a grant from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stijns RCH, Tromp MR, Hugen N, et al. Advances in organ preserving strategies in rectal cancer patients. Eur J Surg Oncol 2018;44:209-19. [Crossref] [PubMed]
- Deng Y. Rectal Cancer in Asian vs. Western Countries: Why the Variation in Incidence? Curr Treat Options Oncol 2017;18:64. [Crossref] [PubMed]
- Gaertner WB, Kwaan MR, Madoff RD, et al. Rectal cancer: An evidence-based update for primary care providers. World J Gastroenterol 2015;21:7659-71. [Crossref] [PubMed]
- Bailey CE, Hu CY, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg 2015;150:17-22. [Crossref] [PubMed]
- Dayde D, Tanaka I, Jain R, et al. Predictive and Prognostic Molecular Biomarkers for Response to Neoadjuvant Chemoradiation in Rectal Cancer. Int J Mol Sci 2017; [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Qadir MI, Faheem A. miRN(A) A Diagnostic and Therapeutic Tool for Pancreatic Cancer. Crit Rev Eukaryot Gene Expr 2017;27:197-204. [Crossref] [PubMed]
- Boyerinas B, Park SM, Hau A, et al. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 2010;17:F19-36. [Crossref] [PubMed]
- Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 2009;174:736-45. [Crossref] [PubMed]
- Shell S, Park S, Radjabi A, et al. Let-7 expression defines two differentiation stages of cancer. PNAS 2007;104:11400-5. [Crossref] [PubMed]
- Ramberg H, Alshbib A, Berge V, et al. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol Cancer 2011;10:50. [Crossref] [PubMed]
- Wei Y, Liu G, Wu B, et al. Let-7d Inhibits Growth and Metastasis in Breast Cancer by Targeting Jab1/Cops5. Cell Physiol Biochem 2018;47:2126-35. [Crossref] [PubMed]
- Jiang J, Liu HL, Tao L, et al. Let7d inhibits colorectal cancer cell proliferation through the CST1/p65 pathway. Int J Oncol 2018;53:781-90. [PubMed]
- Manikandan M, Deva Magendhra Rao AK, Arunkumar G, et al. Oral squamous cell carcinom(A) microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer 2016;15:28. [Crossref] [PubMed]
- Ning YX, Luo X, Xu M, et al. Let-7d increases ovarian cancer cell sensitivity to a genistein analog by targeting c-Myc. Oncotarget 2017;8:74836-45. [Crossref] [PubMed]
- Li Y, Jia Q, Zhang Q, et al. Rab25 upregulation correlates with the proliferation, migration, and invasion of renal cell carcinoma. Biochem Biophys Res Commun 2015;458:745-50. [Crossref] [PubMed]
- Liu K, Zhang C, Li T, et al. Let-7a inhibits growth and migration of breast cancer cells by targeting HMGA1. I Int J Oncol 2015;46:2526. [Crossref] [PubMed]
- Di FR, Drago-Ferrante R, Pentimalli F, et al. Let-7d miRNA Shows Both Antioncogenic and Oncogenic Functions in Osteosarcoma-Derived 3AB-OS Cancer Stem Cells. J Cell Physiol 2016;231:1832-41. [Crossref] [PubMed]
- Su B, Zhao W, Shi B, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer 2014;13:206. [Crossref] [PubMed]
- Wu L, Zhang W, Jia S, et al. Mutation analysis of the ABCC2 gene in Chinese patients with Dubin-Johnson syndrome. Exp Ther Med 2018;16:4201-6. [PubMed]
- Nies AT, König J, Pfannschmidt M, et al. Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer 2001;94:492-9. [Crossref] [PubMed]
- Arts HJ, Katsaros D, de Vries EG, et al. Drug resistance-associated markers P-glycoprotein, multidrug resistance-associated protein 1, multidrug resistance-associated protein 2, and lung resistance protein as prognostic factors in ovarian carcinoma. Clin Cancer Res 1999;5:2798-805. [PubMed]
- Sandusky GE, Mintze KS, Pratt SE, et al. Expression of multidrug resistance-associated protein 2 (MRP2) in normal human tissues and carcinomas using tissue microarrays. Histopathology 2002;41:65-74. [Crossref] [PubMed]
- Tecza K, Pamula-Pilat J, Lanuszewska J, et al. Genetic polymorphisms and response to 5-fluorouracil, doxorubicin and cyclophosphamide chemotherapy in breast cancer patients. Oncotarget 2016;7:66790-808. [Crossref] [PubMed]
- Takegawa N, Nonagase Y, Yonesaka K, et al. DS-8201a, a new HER2-targeting antibody-drug conjugate incorporating a novel DNA topoisomerase I inhibitor, overcomes HER2-positive gastric cancer T-DM1 resistance. Int J Cancer 2017;141:1682-9. [Crossref] [PubMed]
- Andersen V, Svenningsen K, Knudsen LA, et al. Novel understanding of ABC transporters ABCB1/MDR/P-glycoprotein, ABCC2/MRP2, and ABCG2/BCRP in colorectal pathophysiology. World J Gastroenterol 2015;21:11862-76. [Crossref] [PubMed]
- Zhan M, Qiu Q, Wang G, et al. Let-7c sensitizes acquired cisplatin-resistant A549 cells by targeting ABCC2 and Bcl-XL. Pharmazie 2013;68:955-61. [PubMed]
- Surowiak P, Materna V, Kaplenko I, et al. ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of ovarian carcinomas and correlates with resistance to cisplatin and clinical outcome. Clin Cancer Res 2006;12:7149-58. [Crossref] [PubMed]


