
Effects of antisense lncRNA PCBP1-AS1 on biological behaviors of vulvar squamous carcinoma cells by regulating TRAF5 and NF-κB expression
Introduction
Vulvar cancer is one of multiple malignant tumors of the female reproductive organs, the majority of which are primary vulvar squamous cell carcinomas (VSCC) (1). Recently, the incidence of VSCC has increased yearly (2), threatening the health of older women because the five-year survival rate of advanced VSCC is only 20%.
Poly C binding protein-1 antisense RNA1 (PCBP1-AS1) is an antisense long noncoding RNA (lncRNA) of PCBP1. Antisense lncRNA is an endogenous lncRNA molecule that is complementary to the sequence of other transcripts and regulates the expression of target genes at the transcriptional and posttranscriptional levels. Antisense lncRNA may play a unique role in the development of disease. Our group was the first to report a genome-wide expression profile of lncRNA molecules in VSCC tissues compared with corresponding adjacent normal vulvar tissue using microarray analysis technology (3). We found that PCBP1-AS1 and TRAF5 mRNA were significantly coexpressed in VSCC using bioinformatics analysis and target gene prediction (Correlation >0.99, P value <0.05).
Tumor necrosis factor receptor associated factor 5 (TRAF5) is an important intracellular signal transduction protein. Previous studies have shown that TRAF5 mediates activation of the nuclear factor kappa B (NF-κB) signal transduction pathway, playing a regulatory role in the biological behavior of tumor cells (4).
In the study, we aimed to investigate the expression of PCBP1-AS1 in VSCC tissues and analyze its clinical significance. Specifically, we determined that PCBP1-AS1 could affect the proliferation and apoptosis of VSCC SW954 cells through TRAF5-mediated regulation of the NF-κB signaling pathway.
Methods
Tissue samples
Nineteen pairs of VSCC and corresponding adjacent normal vulvar tissues were consecutively collected from patients undergoing surgical resection at the Department of Gynecology, Shengjing Hospital, China Medical University from January 2011 to October 2016. Patients ranged in age from 34 to 85 years, with a mean age of 56.9±13.9 years. All specimens were confirmed by pathological examination, and no patients underwent radiotherapy, chemotherapy, or any other medical intervention prior to surgery. The collected tissue samples were immediately frozen in liquid nitrogen after resection and were stored in liquid nitrogen until RNA and protein extraction. Patients in this study all provided written informed consent. This study was approved by the Ethics Committee of Shengjing Hospital affiliated with China Medical University.
Cell lines and transfection
The SW954 cell line was purchased from Shanghai Huiying Biological Technology co., LTD (Shanghai, China). All cells were cultured in RPMI-1640 medium (HyClone, Logan, Utah, USA) containing 20% heat-inactivated fetal bovine serum (FBS; BioInd, Kibbutz Beit Haemek, Israel) and 1% penicillin/streptomycin (Beyotime Biotechnology, Shanghai, China) in a humidified incubator containing 5% CO2 at 37 °C.
SW954 cells were transfected with either a PCBP1-AS1 overexpression plasmid, a TRAF5 shRNA plasmid, or their corresponding negative control vectors (Genechem, Shanghai, China) individually or simultaneously using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
The cells in the blank control group and TRAF5-RNAi-0 group were treated with recombinant human TNF-alpha (Novus, Colorado, USA), an NF-κB activator, as the activation groups. The cells in the blank control group and PCBP1-AS1-over group were treated with BAY11-7082 (Beyotime, Shanghai, China), an NF-κB inhibitor, as the inhibition groups. The cells in the blank control group, PCBP1-AS1-over group, and TRAF5-RNAi-0 group were left untreated as controls. The concentrations of recombinant human TNF-α and BAY11-7082 were both 2×10–5 g/mL. Cells were collected for further experiments at 24 or 48 h after transfection.
Total RNA extraction, reverse transcription and qRT-PCR
Total RNA was extracted from 19 pairs of VSCC and corresponding adjacent normal vulvar tissue specimens or were cultured cells using Trizol reagent (Takara, Tokyo, Japan). RNA was reverse transcribed into cDNA using a PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Tokyo, Japan). Quantitative RT-PCR was performed using a SYBR® PrimeScriptTM RT-PCR Kit on a Roche LightCycler 480 II real-time PCR platform (Roche, Basel, Switzerland). Gene expression in each sample was normalized to GAPDH expression. The primer sequences used were as follows: for GAPDH, 5'-GCACCGTCAAGGCTGAGAAC-3' (forward) and 5'-TGGTGAAGACGCCAGTGGA-3' (reverse); for PCBP1-AS1, 5'-TGCCAAGAGCCTATCCATTC-3' (forward) and 5'-TCACTCCCTTCACCCTGTCT-3' (reverse); for TRAF5, 5'-AACCTGACCCCAATAGCAGC-3' (forward) and 5'-TCAGTTAAGTCCACGGCCAC-3' (reverse). Real-time PCRs were performed in triplicate. The relative expression levels of PCBP1-AS1 and TRAF5 were calculated using the 2−ΔΔCt value method.
Western blot assay
Total protein samples were collected from tissue specimens and were cultured cells in RIPA lysis buffer (Beyotime, Shanghai, China) or cell lysis buffer (Beyotime, Shanghai, China) containing 1% protease phosphatase inhibitor cocktail (Beyotime, Shanghai, China). Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Massachusetts, USA). The membranes were blocked in Tween-20 containing 5% nonfat milk and were incubated with the following primary antibodies overnight at 4 °C: rabbit anti-human TRAF5 (Proteintech, Chicago, USA; 1:500), rabbit anti-human p65/RELA (Proteintech, Chicago, USA; 1:500), rabbit anti-human phosphorylated NF-κB p65 (pSer536) (Cell Signaling Technology, Boston, USA; 1:500), rabbit anti-human c-Rel (Proteintech, Chicago, USA; 1:500), rabbit anti-human IκBα (Cell Signaling Technology, Boston, USA; 1:500) or mouse anti-human GAPDH (Proteintech, Chicago, USA; 1:10,000). After washing with TBST, the membranes were incubated with either goat anti-rabbit IgG (H + L) (Zhongshan Golden Bridge, Beijing, China; 1:5,000) or goat anti-mouse IgG (Proteintech, Chicago, USA; 1:5,000) secondary antibody diluted with TBST, as appropriate. Proteins were visualized using an electrochemiluminescence (ECL) detection kit (Absin, Shanghai, China), and band intensities were quantified using the Azure c300 Chemiluminescent Image System (Azure Biosystems, Dublin, CA, USA). Quantitative analysis was performed using ImageJ 1.44p software (National Institutes of Health, Bethesda, Maryland, USA). TRAF5, p65, p-p65, c-Rel, and IκBα protein expression levels were normalized to those of GAPDH.
NF-κB activation-nuclear translocation fluorescence labeling assay
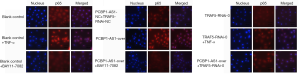
SW954 cells (5×105 cells/well) were seeded into 6-well plates and were transfected with the PCBP1-AS1 overexpression plasmid (PCBP1-AS1-over), TRAF5 shRNA plasmid (TRAF5-RNAi-0), both the PCBP1-AS1 overexpression plasmid and TRAF5 shRNA plasmid (PCBP1-AS1-over + TRAF5-RNAi-0), or the corresponding negative control vectors (PCBP1-AS1-NC + TRAF5-RNAi-NC) for 24 h. The cells were immunofluorescence labeled using a cellular NF-κB Activation-Nuclear Translocation Assay Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol.
Cell proliferation assay
Cell proliferation was evaluated using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan). SW954 cells (2×103 cells/well) were plated in 96-well plates containing 100 µL of complete medium. The cells were incubated overnight and then were transfected with different plasmids. Absorbances were measured at 450 nm using a Synergy H1 Microplate Reader (BioTek, Winooski, Vermont, USA) at each time point. Cell proliferation curves were plotted using the mean absorbance (optical density OD) at each time point. The cell proliferation inhibition rate = [(OD experimental group − OD blank group) − (OD negative control group − OD blank group)]/(OD negative control group − OD blank group) ×100%. The experiment was conducted using both NF-κB activation and inhibition groups as well as control groups.
Cell apoptosis assay
SW954 cells (5×105 cells/well) were seeded into 6-well plates and were transfected with different plasmids. Cells were collected by trypsinization, washed twice with cold PBS, and then were resuspended in 1× Annexin V Binding Buffer at a concentration of 1×106 cells/ml. One hundred microliters of each cell suspension were transferred to a corresponding 5-mL flow tube and was stained with Annexin V-FITC (5 µL) and propidium iodide (5 µL) using the Annexin V-FITC Apoptosis Detection Kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol. The percentage of apoptotic cells was quantified using a BD FACSCalibur Flow Cytometer (BD Biosciences, California, USA).
Wound healing assay
SW954 cells (5×105 cells/well) were seeded into 6-well plates and were transfected with different plasmids. When the cell density reached approximately 90-95% confluence, artificial and homogeneous scratch wounds were created on the confluent monolayer cells using a sterile 200-µL pipette tip. The speed of wound healing was monitored and imaged at three different time points (0, 24, and 48 h) using an Eclipse NI microscope (Nikon, Tokyo, Japan). The percent of wound healing was calculated using ImageJ 1.44p software (National Institutes of Health, Bethesda, Maryland, USA).
Transwell invasion assay
Twenty-four-well Transwell chambers with an 8-µm pore size (Corning, New York, USA) were used to evaluate the cell invasion potential. SW954 cells were transfected with different plasmids. Twenty-four hours after transfection, the cells were harvested. Transfected or blank SW954 cells (2×105 cells/well) were seeded onto Matrigel-coated membranes in the upper chamber containing serum-free RPMI 1640 medium. The bottom chamber contained RPMI 1640 medium with 20% FBS. Cells were incubated at 37 °C with 5% CO2 for 24 h, after which the upper layer of the chamber was scrubbed with a sterile cotton swab to remove any remaining cells, and then the bottom of the chamber was fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Beyotime, Shanghai, China). Cells that invaded through the membrane were counted and photographed in five random fields of each chamber using an Eclipse NI microscope (Nikon, Tokyo, Japan), and then the average number of invasive cells was calculated. Each experiment was performed in triplicate.
Statistical analysis
The data are presented as means ± standard deviation (SD). All statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Differences among data were evaluated using ANOVA for multiple-group comparisons and Student’s t-test for two-group comparisons, whereas the correlations between PCBP1-AS1 or TRAF5 mRNA expression and the clinicopathological characteristics of patients with VSCC were analyzed using Fisher’s exact test. Graph Pad Prism® version 5.01 software (GraphPad Software Inc., San Diego, California, USA) was used to build statistical histograms. Differences were considered statistically significant at a P value <0.05.
Results
The expression of PCBP1-AS1, TRAF5 and NF-κB pathway proteins are downregulated in VSCC tissues
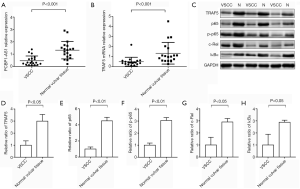
To determine the levels of PCBP1-AS1, TRAF5 and NF-κB pathway proteins in VSCC tissues, qRT-PCR was performed to evaluate the levels of PCBP1-AS1 and TRAF5 mRNA and immunoblotting was used to evaluate the expressions of TRAF5, p65, p-p65, c-Rel, and IκBα protein in 19 paired VSCC tissue samples and their corresponding adjacent normal vulvar tissue. As shown in Figure 1A, the expression of PCBP1-AS1 in VSCC tissues (0.465±0.384) was significantly downregulated compared with the corresponding adjacent normal vulvar tissue (1.321±0.707, P<0.001). As shown in Figure 1B, the expression level of TRAF5 mRNA in VSCC tissues (0.495±0.439) was also markedly decreased compared with the corresponding adjacent normal vulvar tissue (1.336±1.081, P<0.001). Additionally, TRAF5 protein expression in VSCC tissues was significantly lower than normal vulvar tissue (P=0.012) (Figure 1C,D), which was similar with p65 (P<0.01) (Figure 1E), p-p65 (P=0.008) (Figure 1F), c-Rel (P=0.016) (Figure 1G) and IκBα proteins (P=0.032) (Figure 1H).
Correlation between the expression of PCBP1-AS1 and TRAF5 mRNA and clinicopathological characteristics of patients with VSCC
To identify the clinical relevance of PCBP1-AS1 and TRAF5 mRNA expression in VSCC, the correlation between PCBP1-AS1 and TRAF5 mRNA expression and clinicopathological parameters such as age, tumor size, tumor differentiation, FIGO stage, lymph node metastasis, distant metastasis, and squamous cell carcinoma antigen (SCC-Ag) was examined. Using the results of the qRT-PCR assay to establish median values (M) of the relative expression levels of PCBP1-AS1 and TRAF5 mRNA in VSCC tissues as a boundary, we divided the 19 cases into a high expression group and low expression group. The median values were 0.309 for PCBP1-AS1 (M=0.309) and 0.414 for TRAF5 mRNA (M=0.414). P<0.05 was considered statistically significant. As shown in Table 1, PCBP1-AS1 downregulation in VSCC tissues was significantly correlated with the tumor size (P=0.023), tumor differentiation (P=0.020), FIGO stage (P=0.020), lymph node metastasis (P=0.020), distant metastasis (P=0.033) and SCC-Ag (P=0.033) but was not closely related to the patient’s age (P=0.303). Decreased TRAF5 mRNA expression was markedly correlated with the FIGO stage (P=0.020) and lymph node metastasis (P=0.001) but not with other clinical characteristics.
Table 1
| Clinicopathological characteristics | Case number | Expression level of PCBP1-AS1 | Expression level of TRAF5 mRNA | |||||
|---|---|---|---|---|---|---|---|---|
| High | Low | P value | High | Low | P value | |||
| Age/year | 0.303 | 0.582 | ||||||
| ≤50 | 4 | 3 | 1 | 1 | 3 | |||
| >50 | 15 | 6 | 9 | 8 | 7 | |||
| Tumor size (d/cm) | 0.023 | 0.179 | ||||||
| ≤4 | 9 | 7 | 2 | 6 | 3 | |||
| >4 | 10 | 2 | 8 | 3 | 7 | |||
| Tumor differentiation | 0.020 | 0.170 | ||||||
| Well and moderately | 11 | 8 | 3 | 7 | 4 | |||
| Poorly | 8 | 1 | 7 | 2 | 6 | |||
| FIGO stage | 0.020 | 0.020 | ||||||
| I–II | 7 | 6 | 1 | 6 | 1 | |||
| III–IV | 12 | 3 | 9 | 3 | 9 | |||
| Lymph node metastasis | 0.020 | 0.001 | ||||||
| Yes | 12 | 3 | 9 | 2 | 10 | |||
| No | 7 | 6 | 1 | 7 | 0 | |||
| Distant metastasis | 0.033 | 0.303 | ||||||
| Yes | 5 | 0 | 5 | 1 | 4 | |||
| No | 14 | 9 | 5 | 8 | 6 | |||
| SCC-Ag (ìg/L) | 0.033 | 0.582 | ||||||
| ≤4 | 4 | 4 | 0 | 1 | 3 | |||
| >4 | 15 | 5 | 10 | 8 | 7 | |||
PCBP1-AS1, poly C binding protein-1 antisense RNA1; TRAF5, tumor necrosis factor receptor associated factor 5; VSCC, vulvar squamous cell carcinomas.
PCBP1-AS1 regulates the activity of the NF-κB pathway via TRAF5
To evaluate whether PCBP1-AS1 regulates the activity of the NF-κB pathway via TRAF5, the expression of PCBP1-AS1 and TRAF5 in SW954 cells was altered via transfection. The levels of PCBP1-AS1 and TRAF5 mRNA were detected by qRT-PCR, and the expressions of TRAF5 protein and NF-κB pathway proteins, including p65, p-p65, c-Rel, and IκBα protein, were analyzed by immunoblotting. As shown in Figure 2A, compared with the blank control group, PCBP1-AS1 level was markedly increased (P<0.01) in the PCBP1-AS1-over group. Simultaneously, TRAF5 mRNA expression was significantly upregulated (P=0.008). In the TRAF5-RNAi-0 group (Figure 2B), TRAF5 mRNA expression was significantly downregulated (P=0.004), but the PCBP1-AS1 expression level was not markedly altered (P>0.05). In the PCBP1-AS1-over + TRAF5-RNAi-0 group, no obvious change was noted in PCBP1-AS1 expression compared with PCBP1-AS1-over group (P>0.05), whereas the expression of TRAF5 mRNA was significantly higher than TRAF5-RNAi-0 group (P<0.01). Consistently with TRAF5 mRNA alteration, the expressions of TRAF5 and NF-κB pathway proteins were changed in different transfection groups (Figure 2C,D,E,F).
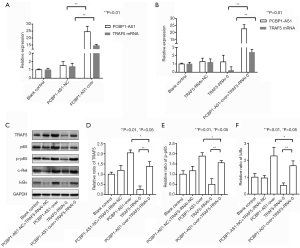
Thus, the results indicated that there were correlation between the expressions of PCBP1-AS1, TRAF5 and NF-κB pathway proteins.
Abnormal expression of PCBP1-AS1 influences SW954 cell proliferation and apoptosis by regulating TRAF5-mediated activation of the NF-κB signaling pathway
To investigate the biological effects of PCBP1-AS1 on VSCC, the experimental groups of SW954 cells with altered expression PCBP1-AS1 and TRAF5 were treated with either an NF-κB signaling pathway activator (TNF-α) or inhibitor (BAY11-7082), and then the CCK-8 assay and Annexin V-FITC & PI staining were conducted to assess cell proliferation and apoptosis, respectively. As shown in Figure 3A, the proliferation of SW954 cells in the PCBP1-AS1-NC + TRAF5-RNAi-NC group was not significantly changed (P>0.05) beginning at 18 h after transfection compared with blank control group. However, cell proliferation in the PCBP1-AS1-over group was significantly decreased (P=0.005), with proliferation inhibition rates of 34.04%, 32.08%, 29.79%, and 28.85% at 18, 24, 36 and 48 h, respectively. Compared with the blank control group, the proliferation of SW954 cells in the TRAF5-RNAi-0 group was markedly accelerated (P=0.005); the cell proliferation inhibition rates at 18, 24, 36, and 48 h were 12.92%, 27.49%, 31.80%, and 28.34%, respectively. Compared with the PCBP1-AS1-over group, the proliferation of SW954 cells in the PCBP1-AS1-over + TRAF5-RNAi-0 group was significantly accelerated (P=0.009) but was drastically decreased compared with the TRAF5-RNAi-0 group (P<0.01). As shown in Figure 3B, compared with the blank control group, the proliferation of SW954 cells in the blank control + TNF-α group was slower (P=0.023), while that of the blank control + BAY11-7082 group was faster (P=0.047). The proliferation of SW954 cells in the PCBP1-AS1-over + BAY11-7082 group was faster than in the PCBP1-AS1-over group (P=0.038). Compared with the TRAF5-RNAi-0 group, the proliferation of SW954 cells in the TRAF5-RNAi-0 + TNF-α was slower (P=0.008).
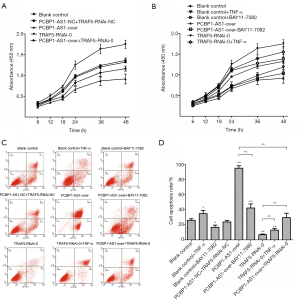
The rate of apoptosis in each experimental group is shown in Figure 3C,D. Compared with the blank control group, the rate of apoptosis cells in the PCBP1-AS1-NC + TRAF5-RNAi-NC group was relatively unchanged while cells in the PCBP1-AS1-over group experienced significantly more apoptosis (P<0.01). Apoptosis was decreased in the TRAF5-RNAi-0 group compared with the blank control (P=0.007). Compared with the PCBP1-AS1-over group, the rate of apoptosis in the PCBP1-AS1-over + TRAF5-RNAi-0 group was drastically decreased (P=0.002) but was increased compared with the TRAF5-RNAi-0 group (P=0.008). Furthermore, compared with the blank control group, the rate of apoptosis in the blank control + TNF-α cells was increased (P=0.046) but was decreased in the blank control + BAY11-7082 cells (P=0.042). The rate of apoptosis in the PCBP1-AS1-over + BAY11-7082 group was markedly lower than that in the PCBP1-AS1-over group (P=0.006). Conversely, apoptosis in the TRAF5-RNAi-0 + TNF-α group was increased compared with that in the TRAF5-RNAi-0 group (P=0.004).
PCBP1-AS1 influences SW954 cell migration and invasion by regulating TRAF5 in an NF-κB-independent manner
To evaluate the effects of PCBP1-AS1 on migration and invasion behavior of VSCC, we performed the wound healing assay and Transwell invasion assay in SW954 cells with altered expression of PCBP1 and TRAF5. As shown in Figure 4A, at 48 hours after in vitro wound induction, SW954 cells in the blank control, blank control + TNF-α, blank control + BAY11-7082, and PCBP1-AS1-NC + TRAF5-RNAi-NC groups had covered approximately 5/8 of the approximately 2-mm wide scratch, whereas cells in the PCBP1-AS1-over and PCBP1-AS1-over + BAY11-7082 groups covered only approximately 1/4 of the scratch. By contrast, cells in the TRAF5-RNAi-0 and TRAF5-RNAi-0 + TNF-α groups had nearly covered all of the scratch. Cells in the PCBP1-AS1-over + TRAF5-RNAi-0 group were covered nearly 1/2 of the scratch.
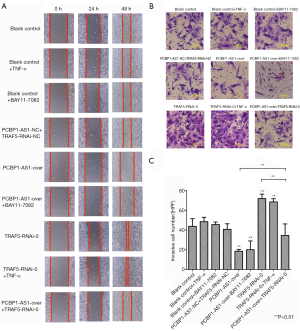
The average number of invasive cells in each experimental group is shown in Figure 4B,C. The Transwell assay was conducted, and the number of invasive cells was counted at the 24-h time point. No significant difference was found in the number of invasive cells in the PCBP1-AS1-NC + TRAF5-RNAi-NC group compared with the blank control group. However, the average number of invasive cells in the PCBP1-AS1-over group was significantly decreased (P=0.006) compared with the blank control group, and the number of invasive cells in the TRAF5-RNAi-0 group was greatly increased (P=0.009) compared with the blank control group. Compared with cells in the PCBP1-AS1-over group, the average number of invasive cells in the PCBP1-AS1-over + TRAF5-RNAi-0 group was increased (P=0.009), but was decreased compared with the number of cells in the TRAF5-RNAi-0 group (P=0.005). Furthermore, compared with the blank control group, no apparent change was noticed in the average number of invasive cells in the blank control + TNF-α group and blank control + BAY11-7082 group (P>0.05). No significant difference was found in the number of invasive cells between the TRAF5-RNAi-0 group and TRAF5-RNAi-0 + TNF-α group (P>0.05). Similarly, no obvious difference was found in the average number of invasive cells in the PCBP1-AS1-over group compared with that in the PCBP1-AS1-over + BAY11-7082 group (P>0.05).
Discussion
Abnormally expressed lncRNA has been identified in various diverse tumor types (5-8). Moreover, abnormal expression of lncRNA is closely related to the proliferative, migratory, invasive, and metastatic abilities of tumor cells and regulation of apoptosis (9-11). However, the expression lncRNAs in VSCC remains to be full elucidated. In our study, we provide evidence that PCBP1-AS1 is aberrantly downregulated and plays a tumor suppressor role in the progression of VSCC. We demonstrated that PCBP1-AS1 positively regulated the expression of TRAF5. Moreover, PCBP1-AS1 exerts the biological effects on SW954 cells via TRAF5-mediated regulation of the NF-κB pathway. Therefore, our study provides a theoretical basis to utilize PCBP1-AS1 in the diagnosis and treatment of VSCC.
Our group has validated the differential expression of HOAIR, MALAT1, MEG3, NEAT1, MIR31HG, and LINC00478 in VSCC and corresponding adjacent normal vulvar tissues (3). Based on the microarray results, we determined that the expression of PCBP1-AS1 was significantly downregulated in VSCC. Presently, only two published reports regarding PCBP1-AS1 are available worldwide. In May 2017, Feng et al. performed transcriptome analysis of oral squamous cell carcinoma compared with healthy oral mucosa; the authors found that PCBP1-AS1 was significantly differentially expressed (12). In the same year, in July, Mo et al. identified a functional lncRNA-mRNA co-expression module between PCBP1-AS1 and target gene mRNA in human peripheral blood mononuclear cells (13). In this study, we measured the relative expression level of PCBP1-AS1 and TRAF5 mRNA in VSCC tissues using qRT-PCR and analyzed the correlation between the expression level of PCBP1-AS1 and TRAF5 mRNA and clinicopathological characteristics of patients with VSCC. Our results indicated that the expression level of PCBP1-AS1 and TRAF5 mRNA in VSCC tissues was significantly downregulated compared with that in corresponding adjacent normal vulvar tissues. PCBP1-AS1 expression was closely related to the tumor size, tumor differentiation, FIGO stage, lymph node metastasis, distant metastasis, and SCC-Ag, while TRAF5 mRNA expression was significantly correlated with the FIGO stage and lymph node metastasis. These results suggest that PCBP1-AS1 and TRAF5 mRNA might be used as indicators for the diagnosis and prognosis of VSCC.
NF-κB, a family of transcription factor proteins, consists of five subunits: c-Rel (Rel), p65 (RelA, NF-κB3), RelB, p50 (NF-κB1), and p52. The five subunits have Rel homology domains and, thus, can form homo- or hetero-dimers, which bind to IκB at rest to form complexes that are present in the cytoplasm in an inactive state. In the classical pathway of activation, after a specific stimulus, IκBα is phosphorylated and ubiquitinated and is degraded by the proteasome; thus, it is dissociated from NF-κB. After being phosphorylated, NF-κB enters the nucleus and binds to a fixed nucleotide sequence in the promoter region of the target gene to regulate the transcription of the target gene (14,15). The NF-κB signaling pathway is thought to play a dual role in the progression of cancer (15-18). Studies have shown that activation of the NF-κB/p65 pathway promotes the progression of breast cancer, colorectal cancer, and head and neck carcinoma (19-21), while the NF-κB/c-Rel pathway is activated in breast and pancreatic cancers (22,23). However, other studies have shown that the NF-κB/p65 pathway is involved in the apoptosis of tumor cells and that the inhibition of the NF-κB/p65 pathway leads to the rapid development of skin cancer (24,25). The NF-κB/c-Rel pathway also plays a tumor suppressor role in lymphomas and gastric carcinomas (26,27). The role of TRAF5-mediated regulation of the NF-κB signaling pathway in inflammation has been widely recognized, and its role in the progression of tumors has also been described (4,28). In our study, these results indicated that the expression of TRAF5, p65, p-p65, c-Rel, and IκBα protein were all significantly downregulated in VSCC tissue. Furthermore, activation of the TRAF5-mediated NF-κB/p65 pathway and NF-κB/c-Rel pathway in normal vulvar tissue and a corresponding decrease in TRAF5 protein expression in VSCC tissue, leading to inhibition of the NF-κB/p65 pathway, with decreased protein expression of p65, IκBα, and p-p65. Additionally, the NF-κB/c-Rel pathway is inhibited in VSCC tissue, resulting in decreased expression of c-Rel protein. Our results demonstrated that TRAF5 mRNA was increased when PCBP1-AS1 expression was upregulated, while PCBP1-AS1 expression was unchanged after TRAF5 expression was downregulated. Upregulation of PCBP1-AS1 significantly increased NF-κB activity, while downregulation of TRAF5 markedly decreased NF-κB activity (Figure S1). Thus, aberrant expression of PCBP1-AS1 can regulate TRAF5 expression both at the level of transcription and translation, and mediate changes in NF-κB pathway activity.
Upregulation of PCBP1-AS1 significantly increased apoptosis and inhibited cell proliferation, migration and invasion; meanwhile, knockdown of TRAF5 significantly inhibited apoptosis and enhanced cell proliferation, migration, and invasion. Moreover, activation or of inhibition the NF-κB signaling pathway suppressed or enhanced proliferation and increased or reduced apoptosis. However, activation and inhibition of the NF-κB signaling pathway had no significant effect on the migration and invasion abilities of the cells. These results suggest that PCBP1-AS1 may regulate the proliferation and apoptosis of VSCC SW954 cells via TRAF5-mediated regulation of the NF-κB signaling pathway, influencing the progression of VSCC, and may play a role in the migration and invasion ability of SW954 cells via an NF-κB signaling pathway-independent mechanism.
Moreover, some limitations exist in our study. First, our sample size was limited, and more VSCC tissues are needed for further study. Second, the detailed molecular mechanism by which PCBP1-AS1 regulated TRAF5 also remains to be fully elucidated. Finally, although PCBP1-AS1 and TRAF5 can regulate the migration and invasion of VSCC, which signaling pathways involved remains unclear in our study.
Conclusions
This study demonstrates that, as a tumor suppressor gene, PCBP1-AS1 plays an important role in the progression of VSCC. PCBP1-AS1 can positively regulate the TRAF5-mediated NF-κB signaling pathway to affect the proliferation and apoptosis of VSCC SW954 cells. Additionally, PCBP1-AS1 can influence the migration and invasion of SW954 cells by regulating TRAF5, but not through the NF-κB signaling pathway.
Acknowledgments
Funding: This study was funded by special research grant from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Patients in this study all provided written informed consent. This study was approved by the Ethics Committee of Shengjing Hospital affiliated with China Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Woelber L, Kock L, Gieseking F, et al. Clinical management of primary vulvar cancer. Eur J Cancer 2011;47:2315-21. [Crossref] [PubMed]
- Clancy AA, Spaans JN, Weberpals JI. The forgotten woman's cancer: vulvar squamous cell carcinoma (VSCC) and a targeted approach to therapy. Ann Oncol 2016;27:1696-705. [Crossref] [PubMed]
- Ni S, Zhao X, Ouyang L. Long non-coding RNA expression profile in vulvar squamous cell carcinoma and its clinical significance. Oncol Rep 2016;36:2571-8. [Crossref] [PubMed]
- Tao T, Cheng C, Ji Y, et al. Numbl inhibits glioma cell migration and invasion by suppressing TRAF5-mediated NF-kappaB activation. Mol Biol Cell 2012;23:2635-44. [Crossref] [PubMed]
- Castillo J, Stueve TR, Marconett CN. Intersecting transcriptomic profiling technologies and long non-coding RNA function in lung adenocarcinoma: discovery, mechanisms, and therapeutic applications. Oncotarget 2017;8:81538-57. [Crossref] [PubMed]
- Collette J, Le Bourhis X, Adriaenssens E. Regulation of Human Breast Cancer by the Long Non-Coding RNA H19. Int J Mol Sci 2017;18: [Crossref] [PubMed]
- Moschovis D, Gazouli M, Tzouvala M, et al. Long non-coding RNA in pancreatic adenocarcinoma and pancreatic neuroendocrine tumors. Ann Gastroenterol 2017;30:622-8. [PubMed]
- Nasrollahzadeh-Khakiani M, Emadi-Baygi M, Nikpour P. Augmented expression levels of lncRNAs ecCEBPA and UCA1 in gastric cancer tissues and their clinical significance. Iran J Basic Med Sci 2017;20:1149-58. [PubMed]
- Eoh KJ, Paek J, Kim SW, et al. Long non-coding RNA, steroid receptor RNA activator (SRA), induces tumor proliferation and invasion through the NOTCH pathway in cervical cancer cell lines. Oncol Rep 2017;38:3481-8. [PubMed]
- Gioia R, Drouin S, Ouimet M, et al. LncRNAs downregulated in childhood acute lymphoblastic leukemia modulate apoptosis, cell migration, and DNA damage response. Oncotarget 2017;8:80645-50. [Crossref] [PubMed]
- Marin-Bejar O, Mas AM, Gonzalez J, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol 2017;18:202. [Crossref] [PubMed]
- Feng L, Houck JR, Lohavanichbutr P, et al. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget 2017;8:31521-31. [PubMed]
- Mo XB, Wu LF, Zhu XW, et al. Identification and evaluation of lncRNA and mRNA integrative modules in human peripheral blood mononuclear cells. Epigenomics 2017;9:943-54. [Crossref] [PubMed]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell 2008;132:344-62. [Crossref] [PubMed]
- Perkins ND. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol 2004;14:64-9. [Crossref] [PubMed]
- Hunter JE, Leslie J, Perkins ND. c-Rel and its many roles in cancer: an old story with new twists. Br J Cancer 2016;114:1-6. [Crossref] [PubMed]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 2006;441:431-6. [Crossref] [PubMed]
- Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer 2012;12:121-32. [Crossref] [PubMed]
- Qin M, Liu S, Li A, et al. NIK- and IKKbeta-binding protein promotes colon cancer metastasis by activating the classical NF-kappaB pathway and MMPs. Tumour Biol 2016;37:5979-90. [Crossref] [PubMed]
- Shafik NM, Mohamed DA, Bedder AE, et al. Significance of Tissue Expression and Serum Levels of Angiopoietin-like Protein 4 in Breast Cancer Progression: Link to NF-kappaB /P65 Activity and Pro-Inflammatory Cytokines. Asian Pac J Cancer Prev 2015;16:8579-87. [Crossref] [PubMed]
- Tamatani T, Azuma M, Aota K, et al. Enhanced IkappaB kinase activity is responsible for the augmented activity of NF-kappaB in human head and neck carcinoma cells. Cancer Lett 2001;171:165-72. [Crossref] [PubMed]
- Chandler NM, Canete JJ, Callery MP. Increased expression of NF-kappa B subunits in human pancreatic cancer cells. J Surg Res 2004;118:9-14. [Crossref] [PubMed]
- Cogswell PC, Guttridge DC, Funkhouser WK, et al. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 2000;19:1123-31. [Crossref] [PubMed]
- Ryan KM, Ernst MK, Rice NR, et al. Role of NF-kappaB in p53-mediated programmed cell death. Nature 2000;404:892-7. [Crossref] [PubMed]
- van Hogerlinden M, Auer G, Toftgard R. Inhibition of Rel/Nuclear Factor-kappaB signaling in skin results in defective DNA damage-induced cell cycle arrest and Ha-ras- and p53-independent tumor development. Oncogene 2002;21:4969-77. [Crossref] [PubMed]
- Burkitt MD, Williams JM, Duckworth CA, et al. Signaling mediated by the NF-kappaB sub-units NF-kappaB1, NF-kappaB2 and c-Rel differentially regulate Helicobacter felis-induced gastric carcinogenesis in C57BL/6 mice. Oncogene 2013;32:5563-73. [Crossref] [PubMed]
- Hunter JE, Butterworth JA, Zhao B, et al. The NF-kappaB subunit c-Rel regulates Bach2 tumour suppressor expression in B-cell lymphoma. Oncogene 2016;35:3476-84. [Crossref] [PubMed]
- Huang S, Wa Q, Pan J, et al. Downregulation of miR-141-3p promotes bone metastasis via activating NF-kappaB signaling in prostate cancer. J Exp Clin Cancer Res 2017;36:173. [Crossref] [PubMed]


