
Circular RNA SMARCA5 is overexpressed and promotes cell proliferation, migration as well as invasion while inhibits cell apoptosis in bladder cancer
Introduction
Bladder cancer is one of the most common malignant cancer globally, with approximately 400,000 new cases each year worldwide, and estimated 272,400 new cases during 2015 in China (1-3). According to tumor stage, bladder cancer could be divided in two major classes: non-muscle-invasive disease (around 70% of all cases) and muscle-invasive bladder cancer (around 30% of all cases) (4,5). Although the majority of patients are diagnosed as non-muscle-invasive bladder cancer, and transurethral resection, as a kind of primarily treatment, has good efficacy on removing tumors, while there are still 50–70% of these tumors are prone to relapse after resection and develop invasive bladder cancer (4,5). Recently, despite various treatments including surgery, chemotherapy and radiotherapy have been applied on bladder cancer, the treatment efficacy is limited, and bladder cancer is still a big challenging problem not only worldwide but also in China (4). Therefore, it is necessary to explore detailed mechanisms involved in the initiation and development of bladder cancer, which might provide new hope for the treatment.
Circular RNA (CircRNA), a class of noncoding RNA family, is characterized by covalently closed loop formed by back-spicing event, and it is commonly named according to the parental genes or particular functions (6-8). Increasing studies have shown that circRNAs are aberrantly expressed in various cancers and they play crucial roles in regulating cancer biological processes (6,7,9). Circular SMARCA5 (circ-SMARCA5), locates on chr4:144464662-144465125, is encoded by SWI/SNF-related matrix-associated actin-dependent regulator of chromatin A5 (SMARCA5) gene (10). Some attentions have been paid on the role of circ-SMARCA5 in cancers, and several previous studies indicate that circ-SMARCA5 serves as an important regulator in these carcinomas such as prostate cancer and hepatocellular carcinoma, whereas no evidence about the function of circ-SMARCA5 on bladder cancer are found (10,11). Thus, we conducted this study to investigate the expression of circ-SMARCA5 in bladder cancer tissues and cells, and further explore the function of circ-SMARCA5 on regulating cell proliferation, apoptosis, migration and invasion in bladder cancer cells, which might provide valuable information for proposing circ-SMARCA5 as therapeutic target in bladder cancer.
Methods
Samples and cells
Ten pairs of human bladder cancer tissue and adjacent tissue were obtained during the surgery. Five human bladder cancer cell lines including TCCSUP, 5637, J82, UM-UC-3 and T-24 were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China), normal human urothelial cell line SV-HUC-1 was purchased from American type culture collection (Manassas, USA). TCCSUP cell line was cultured in 90% Eagle’s Minimum Essential Medium (EMEM) (Invitrogen, USA) and 10% fetal bovine serum (FBS) (Gibco, USA), 5637 and T-24 cell lines were cultured in 90% Roswell Park Memorial Institute (RPMI) 1640 Medium (Sigma, USA) and 10% FBS (Gibco, USA), J82 and UM-UC-3 cell lines were cultured in 90% Minimum Essential Medium (MEM) (Gibco, USA) and 10% FBS (Gibco, USA), and SV-HUC-1 cell line was cultured in 90% Ham’s F-12K (Kaighn’s) Medium (Gibco, USA) and 10% FBS (Gibco, USA). All cells were incubated under 95% air and 5% CO2 at 37 °C. The Ethics Committee of our hospital approved this study before its initiation, and the number of the approval was 20170082. The study was carried out in accordance with the principles expressed in the Declaration of Helsinki. Besides, all subjects provided written informed consents before the initiation of our study.
Measurement of circ-SMARCA5 expression in bladder cancer
Circ-SMARCA5 expression in human bladder cancer tissue and adjacent tissue, as well as in human bladder cancer cell lines and normal human urothelial cell line was measured by quantitative polymerase chain reaction (qPCR).
Transfection
Control overexpression, circ-SMARCA5 overexpression, control shRNA, and circ-SMARCA5 shRNA plasmids were constructed by Shanghai Gene Bio-Tech Company (Shanghai, China) with pCD25-ciR and pGPU6 plasmids. And 0.8 µg of each plasmid was transfected into UM-UC-3 cells as Control(+), Circ-SMARCA5(+), Control(−), and Circ-SMARCA5(−) groups with HilyMax (Dojindo, Japan). Circ-SMARCA5 expression in each group was measured at 24 h after transfection to determine the transfection success or not using qPCR. Additionally, the sequences of circ-SMARCA5 overexpression and circ-SMARCA5 shRNA were as follows: sense sequence, 5'-CACCGAAAAACAAAAGGAGGCCGAAGCCTCCTTTTGTTTTTC-3'; anti-sense sequence, 5'-AAAAGAAAAACAAAAGGAGGCTTCGGCCTCCTTTTGTTTTTC-3'.
Measurement of proliferation, apoptosis, migration and invasion
Cell proliferation ability was measured at 0, 24, 48 and 72 h after transfection using Counting Kit-8 (CCK-8) (Sangon Biotech, China) according to the instruction of manufacturer. Cell apoptosis rate was measured at 48 h by FITC Annexin V Apoptosis Detection Kit II with propidium iodide (AV/PI) (BD, USA) according to the instruction of manufacturer. Apoptotic protein Cleaved Caspase 3 (C-Caspase 3) and anti-apoptotic protein B-cell lymphoma-2 (Bcl-2) were measured at 48 h using Western Blot. Cell migration rate was calculated by dividing migration distance across the gap at 24 h by that at 0 h under microscope (Olympus, Japan) using wound healing assay referring to a previous report (12). In addition, invasive cell count was calculated using Matrigel invasion assay with Matrigel basement membrane matrix (BD, USA) and 8-um Polyvinylidene Fluoride contained transwell filter chamber (Coring, USA) referring to a previous report (12).
qPCR
Circ-SMARCA5 expression was determined by qPCR assay, which was performed as the following steps: (I) extraction of total RNA was performed with TRIzol™ Reagent (Invitrogen, USA); (II) RNase R (Epicentre, USA) was used to digest the linear RNA in each sample of total RNA (1 µg), followed by the agarose gel electrophoresis to check the residual linear RNA; (III) if no band of linear RNA SMARCA5 was observed, the reverse transcription to cDNA was performed by PrimeScript™ RT reagent Kit (Takara, Japan); (IV) qPCR was conducted by PrimeScript™ RT reagent Kit(TAKARA, Japan), and qPCR amplification was performed at 95 °C for 3 mins, followed by 40 cycles of 95 °C for 5 s, 61 °C for 30 s, and then 72 °C for 30 s. The results of qPCR were calculated by 2-ΔΔCt formula. Notably, we chose the forward and reverse primers according to the unique structure of circular RNA, and the sequences amplified by our primers would contain the sequence that expands the back-splicing junction in circular RNA structure, which was not included in linear RNA. Thus, only the circular RNA was detected in our study. Meanwhile, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference and its qPCR was performed with total RNA. Additionally, the SMARCA5 expression was also detected by qPCR assay. Information of qPCR primers were as follows: Circ-SMARCA5 (hsa_circ_0001445), forward (5'-3'): AGATGGGCGAAAGTTCACTTAGA, reverse (5'-3'): GATTCTGATCCACAAGCCTCCT. GAPDH, forward (5'-3'): GACCACAGTCCATGCCATCAC, reverse (5'-3'): ACGCCTGCTTCACCACCTT. SMARCA5, forward (5'-3'): CCGAGGATTAAACTGGCTCA, reverse (5'-3'): GAGGCCCAGGAATGTTTCTA.
Western Blot
Western Blot assay was performed as the following steps: (I) RIPA Lysis and Extraction Buffer (Thermo, USA) was added to cell samples on ice, and concentration of total protein was assessed by bicinchoninic acid (BCA) kit (Pierce Biotechnology, USA); (II) thermal denaturation was conducted at 98 °C for 5 min; (III) 20 µg proteins were added to the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Thermo, USA); (IV) after electrophoresis, proteins were transferred to polyvinylidene fluoride membranes (PVDF) membrane (Millipore, USA); (V) membranes were blocked with 5% bovine serum albumin (BSA) (Sigma, USA) (37 °C, 1 h), and then incubated with primary antibody (4 °C, overnight) and appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (room temperature, 2 h). Finally, the bands were visualized by Pierce™ECL Plus Western Blotting Substrate (Thermo, USA). Information of antibodies used were as follows: Primary antibody: Caspase-3 Rabbit mAb (CST, USA) (dilution: 1:1,000); Cleaved Caspase-3 Rabbit mAb (CST, USA) (dilution: 1:1,000); Bcl-2 Rabbit mAb (CST, USA) (dilution: 1:1,000); GAPDH Rabbit mAb (CST, USA) (dilution: 1:1,000); Secondary Antibody: Goat Anti-Rabbit IgG H&L (HRP) (CST, USA) (dilution: 1:4,000).
Statistics
Statistical analysis and graphs were conducted using SPSS 22.0 Software (IBM, USA) and GraphPad 6.01 Software (GraphPad Int., USA). Data were presented as mean ± standard deviation, comparison among groups was determined by One-way ANOVA followed by Dunnett’s multiple comparison test, comparison between two groups was determined by t test. P<0.05 was considered as significant.
Results
Circ-SMARCA5 expression in tissue samples and cells
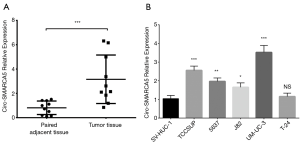
Circ-SMARCA5 expression was remarkably higher in tumor tissue than that in paired adjacent tissue (P<0.001) (Figure 1A). In cell lines, circ-SMARCA5 expression was elevated in various bladder cancer cells including TCCSUP (P<0.001), 5637 (P<0.01), J82 (P<0.05) and UM-UC-3 (P<0.001) cells compared to normal human urothelial cell line SV-HUC-1, while no difference of circ-SMARCA5 expression was found between T-24 cells and SV-HUC-1 cells (P>0.05) (Figure 1B).
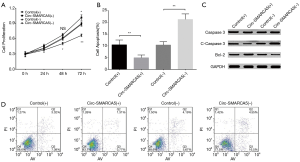
Effect of circ-SMARCA5 on cell proliferation and cell apoptosis in UM-UC-3 cells
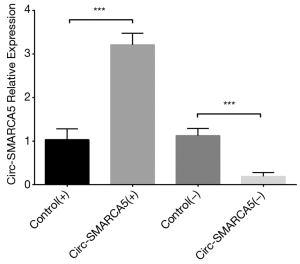
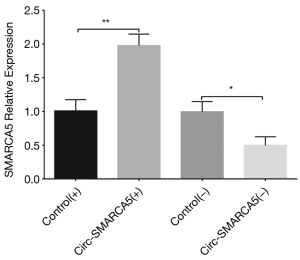
In order to evaluate the function of circ-SMARCA5 in bladder cancer, we performed upregulation and downregulation of circ-SMARCA5 by transfection with corresponding plasmids into UM-UC-3 cells. After 24 h after transfection, circ-SMARCA5 expression was increased in Circ-SMARCA5(+) group compared to Control(+) group (P<0.001), while it was decreased in Circ-SMARCA5(−) group compared to Control(−) group (P<0.001), indicating the plasmids were successfully transfected (Figure 2). Moreover, cell proliferation was raised in Circ-SMARCA5(+) group compared to Control(+) group at 72 h (P<0.05), while it was reduced in Circ-SMARCA5(−) group compared to Control(−) group at 48 h (P<0.05) and 72 h (P<0.01) (Figure 3A). Regarding the cell apoptosis rate at 48 h, it was lower in Circ-SMARCA5(+) group than that in Control(+) group (P<0.01) but higher in Circ-SMARCA5(−) group compared to Control(−) group (P<0.01) (Figure 3B). Meanwhile, expressions of apoptosis-related proteins were detected by Western Blot assay, which displayed that apoptotic protein C-Caspase 3 expression was decreased in Circ-SMARCA5(+) group but increased in Circ-SMARCA5(−) group compared to the corresponding groups, while anti-apoptotic protein Bcl-2 expression was elevated in Circ-SMARCA5(+) group but reduced in Circ-SMARCA5(−) group compared to the corresponding groups (Figure 3C). Besides, the results displayed by Flow Cytometry also showed that cell apoptosis rate was decreased in Circ-SMARCA5(+) group than that in Control(+) group but increased in Circ-SMARCA5(−) group compared to Control(−) group at 48 h (Figure 3D). These results indicated that circ-SMARCA5 enhanced cell proliferation but repressed cell apoptosis in UM-UC-3 cells. Additionally, we found that SMARCA5 expression was elevated in Circ-SMARCA5(+) group compared to Control(+) group (P<0.01) but decreased in Circ-SMARCA5(−) group compared to Control(−) group (P<0.05) (Figure 4).
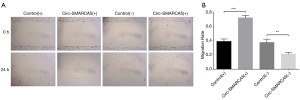
Effect of circ-SMARCA5 on cell migration in UM-UC-3 cells
Cell migration was measured by wound healing assay, which showed that migration rate was raised in Circ-SMARCA5(+) group compared to Control(+) group (P<0.001), while it was reduced in Circ-SMARCA5(−) group compared to Control(−) group (P<0.01) (Figure 5). These data indicated that circ-SMARCA5 promoted cell migration in UM-UC-3 cells.
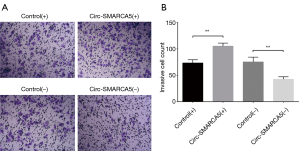
Effect of circ-SMARCA5 on cell invasion in UM-UC-3 cells
Cell invasion was detected by Matrigel invasion assay, and we found increased invasive cell count in the Circ-SMARCA5(+) group compared to Control(+) group (P<0.01) and decreased invasive cell count in the Circ-SMARCA5(−) group compared to Control(−) group (P<0.01). These data suggested that circ-SMARCA5 enhanced cell invasion in UM-UC-3 cells (Figure 6).
Discussion
In this study, we found that circ-SMARCA5 was overexpressed in tumor tissues and tumor cells compared to the paired adjacent tissues and human urothelial cell line respectively. Furthermore, in vitro experiments displayed that circ-SMARCA5 promoted cell proliferation, repressed cell apoptosis, facilitates cell migration and enhanced cell invasion in UM-UC-3 cells.
CircRNAs, are recently recognized as a class of abundant endogenous non-coding RNAs with great regulatory potency in human mammalian cells owing to the development of next generation sequencing technology and bioinformatics. CircRNAs possess some prominent features as follows: (I) the stability due to the covalently closed loop structure (8,13); (II) the universality in human cells (14,15); (III) the specificity varying in tissues and developmental stages (13,16); (IV) the conservatism in different species (17). These features make circRNAs serve as potential biomarkers for human diseases (8,13,14,17-20). Furthermore, according to previous data, some major functions of circRNAs are as follows: (I) acting as miRNA sponges: circRNAs, which have many miRNA response elements, competitively bind to miRNAs and result in decrease of the functional miRNAs (8,13,21); (II) regulators of parental gene transcription: circ-RNAs are able to regulate the transcription of their parental genes (8); (III) circRNA-protein interaction: circRNAs could interact with proteins, and the interaction contributes to regulating transcription of parental genes, promoting interaction of multiple proteins, and altering subcellular localization of proteins (22). Based on these understandings of circRNAs, the investigation of circRNAs in cancer pathology and development has become a hotspot in the field of cancer research, including bladder cancer (8). For instance, an investigation using microarray assay finds 469 dysregulated circRNAs in bladder cancer, among which 285 were up-regulated (9). Moreover, a previous study shows that circular RNA Centrosomal protein 128 (circ-CEP128) enhances cell proliferation and represses cell apoptosis in bladder cancer through upregulating SRY-box-containing gene 11 (SOX11) (6). Also, an interesting study displays that circ_0000144 promotes cell proliferation and invasion through inhibiting miR-217 and upregulating runt-related transcription factor 2 (RUNX2) in bladder cancer cells (7). And another study discloses that circRNA Vang-like protein 1 (circ-VANGL1) is overexpressed in tumor tissues compared to adjacent normal tissues of bladder cancer, moreover, its promotive effects on cell proliferation, migration and invasion via suppressing miR-605-3p have been observed in in vitro assays (23). These previous data have revealed that circRNAs may serve as crucial regulators in the initiation and progression of bladder cancer through affecting the biological processes such as cell proliferation, apoptosis, migration and invasion.
Circ-SMARCA5, the circRNA deriving form exons 15 and 16 of SMARCA5 gene, have been reported to be involved in the pathology of some cancers (10,11). A previous study discloses that circ-SMARCA5 is upregulated in prostate cancer tissues compared to normal prostate tissues and its silencing inhibits prostate cancer cells proliferation but promotes cell apoptosis (10). As to the detailed mechanisms of how circ-SMARCA5 affects tumor cell apoptosis, only a previous study shows that downregulation of SMARCA5, which is the upstream gene of circ-SMARCA5, causes deficiencies in DNA repair and consequently promotes chromosomal instability, and then leads to increased cell apoptosis in bladder cancer. Meanwhile, downregulation of circ-SMARCA5 decreased SMARCA5 expression in our study. Taken together, we speculated that the influence on DNA repair and chromosomal instability might also underlay the mechanisms of circ-SMARCA5 in cell apoptosis, while the direct evidence of circ-SMARCA5 in affecting cell apoptosis was not reported (24). Whereas, another study reveals that circ-SMARCA5 is downregulated in hepatocellular carcinoma tissues compared to adjacent noncancerous liver tissues, moreover, circ-SMARCA5 protects tumor suppressors through the miR-17-3p/miR-181b-5p-TIMP3 pathway, thereby represses cell proliferation in hepatocellular carcinoma cells (11). Although these previous studies show different roles of circ-SMARCA5 in different cancers, they emphasize that circ-SMARCA5 may be a crucial regulator in the pathology of cancers. However, limited evidences reveal the function of circ-SMARCA5 in bladder cancer. Considering circ-SMARCA5 has shown critical regulatory influence in several carcinomas, we supposed that circ-SMARCA5 might also have effects in bladder cancer. To prove our hypothesis, in this study, we firstly compared the circ-SMARCA5 expression between tumor tissue and the paired adjacent tissue, as well as cancer cell lines and normal human urothelial cell line, which showed that circ-SMARCA5 was overexpressed in tumor tissue as well as tumor cells. Subsequently, we conducted several in vitro experiments including CCK-8, AV/PI, wound healing and Matrigel invasion assays, and we observed that circ-SMARCA5 promoted cell proliferation but repressed cell apoptosis in UM-UC-3 cells, furthermore, circ-SMARCA5 facilitated the migration and invasion abilities of UM-UC-3 cells. The possible reasons of these results were that: (I) circ-SMARCA5 might regulate SMARCA5, which is important for gene expression, DNA replication, DNA repair and the maintenance of chromatin structure, thereby affects the biological processes such as proliferation, apoptosis, migration and invasion of bladder cancer cells (25-27); (II) circ-SMARCA5 might sponge some target miRNAs to affect the proliferation, apoptosis, migration and invasion of bladder cells, while the related data of its target miRNAs was limited. Despite further study was needed to enrich the knowledge on circ-SMARCA5 in bladder cancer cells, these results provided supports to understand the mechanisms of circ-SMARCA5 in bladder cancer and shed a light on the application of circ-SMARCA5 as a treatment target in this disease. In addition, there were some limitations existed in our study: (I) changes in cellular morphology induced by manipulating circ-SMARCA5 was not recorded, and further study investigating the effect of circ-SMARCA5 on EMT was needed, which would give important clues for the role of circ-SMARCA5 in malignant process; (II) since this study was not intended to do a detailed mechanistic study, circ-SMARCA5-mediated mechanisms of cell apoptosis, migration and invasion were not investigated. According to previous data, the possible reason for the influence of circ-SMARCA5 on cell activities might be account of that circ-SMARCA5 regulated some target genes (such as miR-103 or miR-202) to affect the cell apoptosis, migration and invasion, while further mechanistic study was needed; (III) it was better to validate the knock-down and overexpression results in another cell line, while this was a preliminary study to investigate the function of circ-SMARCA5 on cell activities, and further study validating these results with more cell lines was needed.
In conclusion, circ-SMARCA5 is overexpressed in bladder cancer tissues and cell lines, and it promotes cell proliferation, migration and invasion, but represses apoptosis in bladder cancer cells.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee of our hospital approved this study before its initiation, and the number of the approval was 20170082. The study was carried out in accordance with the principles expressed in the Declaration of Helsinki (as revised in 2013). Besides, all subjects provided written informed consents before the initiation of our study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Inamura K. Bladder Cancer: New Insights into Its Molecular Pathology. Cancers (Basel) 2018;10: [Crossref] [PubMed]
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Ma G, Yang X, Liang Y, et al. Precision medicine and bladder cancer heterogeneity. Bull Cancer 2018;105:925-31. [Crossref] [PubMed]
- Editorial Office I. International Journal of Molecular Science 2018 Best Paper Award. Int J Mol Sci 2018;19: [Crossref]
- Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med 2018;24:40. [Crossref] [PubMed]
- Huang W, Lu Y, Wang F, et al. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene 2018;678:337-42. [Crossref] [PubMed]
- Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267-74. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Kong Z, Wan X, Zhang Y, et al. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun 2017;493:1217-23. [Crossref] [PubMed]
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. [Crossref] [PubMed]
- Huang K, Geng J, Wang J. Long non-coding RNA RP11-552M11.4 promotes cells proliferation, migration and invasion by targeting BRCA2 in ovarian cancer. Cancer Sci 2018;109:1428-46. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Wang D, Yang S, Wang H, et al. The progress of circular RNAs in various tumors. Am J Transl Res 2018;10:1571-82. [PubMed]
- Yang X, Yuan W, Tao J, et al. Identification of circular RNA signature in bladder cancer. J Cancer 2017;8:3456-63. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Cui X, Wang J, Guo Z, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 2018;17:123. [Crossref] [PubMed]
- Zeng Z, Zhou W, Duan L, et al. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol 2019;234:3887-96. [Crossref] [PubMed]
- Morais DR, Reis ST, Viana N, et al. The involvement of miR-100 in bladder urothelial carcinogenesis changing the expression levels of mRNA and proteins of genes related to cell proliferation, survival, apoptosis and chromosomal stability. Cancer Cell Int 2014;14:119. [Crossref] [PubMed]
- Gigek CO, Lisboa LC, Leal MF, et al. SMARCA5 methylation and expression in gastric cancer. Cancer Invest 2011;29:162-6. [Crossref] [PubMed]
- Delto JC, Paulucci D, Helbig MW, et al. Robot-assisted partial nephrectomy for large renal masses: a multi-institutional series. BJU Int 2018;121:908-15. [Crossref] [PubMed]
- Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A 2003;100:14097-102. [Crossref] [PubMed]


