
Clinic-pathologic features and gene fusion pattern of ALK and ROS1 in non-small cell lung cancer show association with household coal combustion
Introduction
Lung cancer has been the most common cancer for decades (1). Globally about 53% of lung cancer cases in females and 15% of lung cancer cases in males are not attributed to smoking (2). Nearly 3 billion people on earth use coal or biomass for cooking and heating (3,4), this practice poses long-term risks for the development of cardiovascular and respiratory diseases including lung cancer (3-6). Evidences suggest that lung cancer caused by non-tobacco factors can harbor unique clinical features when compared with lung cancers attributed to smoking (2,7,8).
Previous investigations on different populations have identified remarkable diversity in non-smoking lung cancer patients, including epidemiological, clinical and molecular characteristics, furthermore, highlighted fuel choice as an important factor in lung cancer etiology (2,6-9). Interestingly, nonsmokers in certain regions of China experience some of the highest lung cancer rates in the world (5,7,10). The rural counties like Xuanwei and Fuyuan in China’s Yunnan Province have been a particular focus in large-scale epidemiological studies, due to the notably high lung cancer rate among non-smoking female residents (5,7), which have been partially attributed to indoor coal combustion from cooking and heating (4,6).
Driver mutations rise in genes that encode signal molecules which play key roles in the regulation of cell death and proliferation (7). There is a spectrum of driver mutations in lung cancer (EGFR, KRAS, BRAF, HER2, NRAS, PIK3CA, MEK1, AKT1, and PTEN). Other than point mutations, fusion genes which require chromosome rearrangements often represent bigger genomic events and pose a unique signature in non-small cell lung cancer (NSCLC) patients (7,8,11). Previous reports suggest that lung cancer patients who are nonsmokers but coal users show unique driver mutation patterns (7,9). Here is the hypothesis that NSCLC patients who use coal for indoor cooking and heating are exposed to a particular spectrum of carcinogens, thus may possibly harbor unique fusion gene patterns in their tumors.
Despite the abundance of studies performed in this area to date, the net interaction between environmental carcinogens and population genetic background remain complex and dynamic (8,11). For this reason, present study sought to investigate gene rearrangement events in patients exposed to household coal combustion, including ALK, ROS1, RET and NTRK1. Theoretically, reverse transcriptase-polymerase chain reaction (RT-PCR), fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) are three standard methods for the detection of fusion genes (11-14). RT-PCR could detect not only the presence of fusion genes and also the variant types, and it is suitable for a fast, sensitive and high-flux investigation.
Methods
Patients and tissues
This trial was designed as a single-center observational study. Patients were recruited from Department of Thoracic Surgery I of Yunnan Cancer Hospital between 2015 and 2017. To investigate indoor air pollution caused by household coal use (HCU), subjects were selected by the following criteria: (I) the subject can be a resident of Xuanwei and Fuyuan region or other rural areas in Yunnan Province; (II) positive subject uses coal predominantly for cooking and heating in home, and has lived in that coal-using condition for at least 10 years; (III) negative subject reported no history of occupational or domestic coal use; (IV) all the subjects didn’t receive radiotherapy or ALK inhibitor before surgery. The smoking history and coal-using history were obtained based on self-report during patient interview and confirmed by personal medical records. In total 205 subjects were enrolled and 112 were domestic coal users.
Clinic-pathological data were documented in hospital cooperated databank (https://www.linkdoc.com). The stage was reviewed according to the 8th edition of The International Association for the Study of Lung Cancer (IASLC) staging system. The tissue samples were stored in liquid nitrogen at the time of resection. A slide was cut from each sample for HE stain, those containing >70% cancer cells and <10% necrosis was enrolled. The ethical committees of Yunnan Cancer Hospital approved the study (No.KY2019.57). All patients provided informed consent.
RNA extraction and Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed using Promega reverse transcription kit (Promega, Madison, WI, USA), complementary DNA (cDNA) was used for further PCR reactions (Sigma, St. Louis, MO, USA). PCR products were first examined via electrophoresis, and those with right size were sequenced in BGI Tech, Shenzhen, China (http://www.bgitechsolutions.com). The sequences were analyzed via NCBI-BLAST for fusion genes. All positive fusion cases were confirmed by another independent PCR reaction. The PCR conditions were provided in the supplemental material and Table S1.
Compare with other studies
Eighteen studies were summarized, which used RT-PCR for detecting ALK fusions, and provided subjects’ ethnicity and variant type data for EML4-ALK fusion gene. In total, 3,535 NSCLC patients containing 236 ALK fusion cases were included. We further divided the 13 Asian studies based on gender rate and smoking rate of ALK+ subjects (patients positive for ALK fusions) for detailed comparison.
Statistical analysis
Chi-square test and Fischer’s exact test were used to analyze the association, using SPSS 17.0 (SPSS Institute, Chicago, IL, USA). Statistical significance was set at P<0.05 (two-sided P value).
Results
Clinic-pathological features of subjects
The 205 subjects came from Xuanwei and Fuyuan region or other rural areas of Yunnan Province. The clinic-pathological characteristics were shown in Table 1 and Figure 1. The male/female ratio was 1.3/1 (116/89), and the average age was 57 years (range, 38–82 years); 159 (77.6%) subjects were diagnosed as adenocarcinoma (AD), 46 (22.4%) with squamous cell carcinoma (SCC). There were 112 (54.6%, male: 69, female: 43) domestic coal users and 96 (46.8%, all males) smokers. In order to better reflect air carcinogen exposure, both factors were evaluated individually and collectively (Table 2), the union set of smokers and coal users included 145 (70.7%) patients, and 63 (30.7%) were double-positive for smoking and coal-using.
Table 1
| Variables | NSCLC patients (n=205) |
|---|---|
| Average age (years) | 57 (range, 38–82) |
| Gender, n (%) | |
| Male | 116 (56.6) |
| Female | 89 (43.4) |
| Histology, n (%) | |
| Adenocarcinoma | 159 (77.6) |
| Squamous carcinoma | 46 (22.4) |
| Tumor stage, n (%) | |
| I | 98 (47.8) |
| II | 43 (21.0) |
| IIIA | 52 (25.4) |
| IIIB–IV | 12 (5.9) |
| Ex- or current smoker, n (%) | |
| Yes | 96 (46.8) |
| No | 109 (53.2) |
| HCU, n (%) | |
| Yes | 112 (54.6) |
| No | 93 (45.4) |
| ALK rearrangement, n (%) | |
| EML4-ALK | 18 (8.8) |
| Negative | 187 (91.2) |
| ROS rearrangement, n (%) | |
| CD74-ROS1 | 5 (2.4) |
| Negative | 200 (97.6) |
NSCLC, non-small cell lung cancer.
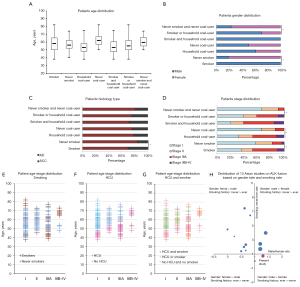
Table 2
| Variables | Smoker (n=96) | P | HCU (n=112) | P | Smoker or HCU (n=145) | P | Smoker and HCU (n=63) | P |
|---|---|---|---|---|---|---|---|---|
| Average age (years) | 58 | 54 | 56 | 54 | ||||
| Gender, n (%) | 5.6E-32 | 0.11 | 6.5E-10 | 6.6E-17 | ||||
| Male | 96 (82.8) | 69 (59.5) | 102 (87.9) | 63 (54.3) | ||||
| Female | 0 (0.0) | 43 (48.3) | 43 (48.3) | 0 (0.0) | ||||
| Histology, n (%) | 0.012 | 0.70 | 0.37 | 0.16 | ||||
| Adenocarcinoma | 67 (42.1) | 88 (55.3) | 110 (69.2) | 45 (28.3) | ||||
| Squamous carcinoma | 29 (63.0) | 24 (52.2) | 35 (76.1) | 18 (39.1) | ||||
| Tumor stage, n (%) | – | – | – | – | ||||
| I | 35 (35.7) | 36 (36.7) | 58 (59.2) | 13 (13.3) | ||||
| II | 16 (37.2) | 24 (55.8) | 28 (65.1) | 12 (27.9) | ||||
| IIIA | 36 (69.2) | 40 (76.9) | 47 (90.4) | 29 (55.8) | ||||
| IIIB–IV | 9 (75.0) | 12 (100.0) | 12 (100.0) | 9 (75.0) | ||||
| ALK fusion, n (%) | 0.44 | 0.048 | 0.027 | 0.43 | ||||
| EML4-ALK | 10 (55.6) | 14 (77.8) | 17 (94.4) | 7 (38.9) | ||||
| Negative | 86 (46.0) | 98 (52.4) | 128 (68.4) | 56 (29.9) | ||||
| ROS fusion, n (%) | 0.37 | 0.065 | 0.32 | 1.0 | ||||
| CD74-ROS1 | 1 (20.0) | 5 (100.0) | 5 (100.0) | 1 (20.0) | ||||
| Negative | 95 (47.5) | 107 (53.5) | 140 (70.0) | 62 (31.0) |
The percentage is calculated based on the total number of each variable listed on the left side. P value behind each subgroup is calculated between this subgroup and its complementary set. P value calculated by chi-square test or Fisher’s exact test, when there is at least a cell frequency less than 5; % is the ratio in certain type of variables; Smoker or HCU means union set of smoker and coal user; Smoker and HCU means double-positive subjects, being a smoker and coal user at the same time. NSCLC, non-small cell lung cancer; HCU, household coal user.
The patients age distribution revealed interesting patterns (Figure 1A). Although no apparent age difference was observed between smokers and nonsmokers, HCU developed lung cancer much earlier than never coal users (average age: 54 vs. 62 years). On the other hand, if tobacco and coal exposure were combined, subjects in union set were much younger than double-negative subjects (average age: 56 vs. 60 years), and double-positive patients were even 2 years younger than union set patients (average age 54 years). There was no apparent gender imbalance on coal use, but the smoking rate difference between two genders was big: nonsmokers were only 17% in males versus 100% in females, so the union set had more males, while the double-negative group included more females (Figure 1B). Adenocarcinoma was the dominate histology in all subgroups, only smokers had statistically more SCC (Figure 1C). For stage at diagnosis, there were more stage IIIA and IIIB–IV cases in smokers and HCU patients. Combining both factors, patients in union set also had increased stage IIIA–IV case (59.31%), if double-positive subjects were selected out, 60.32% of them were in stage IIIA–IV (Figure 1D).
When age, stage and exposure were considered together, increased smokers were found in stage IIIA–IV, but there was only small age distribution imbalance (Figure 1E). HCU patients were apparently younger in all the stages and had a higher ratio in stage IIIA–IV (Figure 1F). Double-negative subjects were mainly in stage I–II with relatively older age. Union set patients covered large age span (40–82 years), showed clearly early-onset and made the majority of stage IIIA–IV cases. Double-positive patients were mainly found in stage IIIA–IV with wider age span (range, 38–75 years) (Figure 1G).
ALK rearrangement
Among the 205 patients, eighteen patients (18/205, 8.8%), ten males and eight females were found to have ALK rearrangement (Table 3). Patients with ALK fusions tended to be younger, with 11 less than 55 years and 7 more than 55. All 18 patients had EML4-ALK fusion gene and they all had adenocarcinoma (P=0.015). In the 18 EML4-ALK cases, 4 variant 1 and 14 variant 3a+3b were identified (Table S2). ALK rearrangement showed no significant association with gender, age, and smoking, but it was associated with household coal combustion (P=0.048), 14 patients were current or ex-coal users and only 4 patients were never coal users. When smoking and coal-using were combined (union set, n=145), P value was improved (P=0.027). Seventeen patients were either coal users or smokers, and only 1 was double-negative. Regarding to stage, most of the patients with ALK rearrangement were in stage I (10/18, 55.6%) and stage IIIA (6/18, 33.3%).
Table 3
| Variables | ALK rearrangements, n (%) | ROS1 rearrangements, n (%) | |||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | P | Positive | Negative | P | ||
| Total number of patients | 18 (8.8) | 187 (91.2) | 5 (2.4) | 200 (97.6) | |||
| Gender | 0.93 | 0.17 | |||||
| Male | 10 (8.6) | 106 (91.4) | 1 (0.9) | 115 (99.1) | |||
| Female | 8 (9.0) | 81 (91.0) | 4 (4.5) | 85 (95.5) | |||
| Age, years | 0.062 | 0.16 | |||||
| <55 | 11 (13.3) | 72 (86.7) | 4 (4.8) | 79 (95.2) | |||
| ≥55 | 7 (5.7) | 115 (94.3) | 1 (0.8) | 121 (99.2) | |||
| Histology | 0.015 | 0.59 | |||||
| Adenocarcinoma | 18 (11.3) | 141 (88.7) | 5 (3.1) | 154 (96.9) | |||
| Squamous cell carcinoma | 0 (0.0) | 46 (100.0) | 0 (0.0) | 46 (100.0) | |||
| Stage | 0.42 | 0.58 | |||||
| I | 10 (10.2) | 88 (89.8) | 3 (3.1) | 95 (96.9) | |||
| II | 2 (4.7) | 41 (95.3) | 0 (0.0) | 43 (100.0) | |||
| IIIA | 6 (11.5) | 46 (88.5) | 2 (3.8) | 50 (96.2) | |||
| IIIB–IV | 0 (0.0) | 12 (100.0) | 0 (0.0) | 12 (100.0) | |||
| Smoking history | 0.44 | 0.37 | |||||
| Yes (current or ex-smoker) | 10 (10.4) | 86 (89.6) | 1 (1.0) | 95 (99.0) | |||
| Never | 8 (7.3) | 101 (92.7) | 4 (3.7) | 105 (96.3) | |||
| Household coal use | 0.048 | 0.065 | |||||
| Yes (current or ex-user) | 14 (12.5) | 98 (87.5) | 5 (4.5) | 107 (95.5) | |||
| Never | 4 (4.3) | 89 (95.7) | 0 (0.0) | 93 (100.0) | |||
| Overall exposure (tobacco and coal) | 0.027 | 0.32 | |||||
| Smoker or household coal-user | 17 (11.7) | 128 (88.3) | 5 (3.4) | 140 (96.6) | |||
| Never smoker & never coal-user | 1 (1.7) | 59 (98.3) | 0 (0.0) | 60 (100.0) | |||
The percentage is calculated based on the total number of each variable listed on the left side. P value calculated by chi-square test or Fisher’s exact test, when there is at least a cell frequency less than 5; smoker or household coal-user: union set of smoker and coal user; never smoker & never coal-user: double-negative patient, not a smoker and also not a coal user. NSCLC, non-small cell lung cancer; RT-PCR, reverse transcriptase polymerase chain reaction; ALK, anaplastic lymphoma kinase; ROS1, ROS proto-oncogene 1.
Table S2
| No. | Gender | Age | Pathology diagnosis | Stage | Smoking history | Household coal combustion | ALK fusion genes detected by RT-PCR |
|---|---|---|---|---|---|---|---|
| 1 | F | 46 | AD | I | N | Y | EML4-ALK V1 |
| 2 | M | 43 | AD | I | Y | Y | EML4-ALK V1 |
| 3 | M | 61 | AD | IIIA | Y | N | EML4-ALK V1 |
| 4 | M | 54 | AD | II | Y | Y | EML4-ALK V1 |
| 5 | F | 53 | AD | I | N | Y | EML4-ALK V3a + V3b |
| 6 | M | 47 | AD | IIIA | Y | Y | EML4-ALK V3a + V3b |
| 7 | F | 57 | AD | II | N | Y | EML4-ALK V3a + V3b |
| 8 | F | 60 | AD | IIIA | N | Y | EML4-ALK V3a + V3b |
| 9 | M | 64 | AD | IIIA | Y | N | EML4-ALK V3a + V3b |
| 10 | M | 44 | AD | I | Y | Y | EML4-ALK V3a + V3b |
| 11 | F | 50 | AD | I | N | Y | EML4-ALK V3a + V3b |
| 12 | M | 53 | AD | I | Y | Y | EML4-ALK V3a + V3b |
| 13 | F | 45 | AD | I | N | Y | EML4-ALK V3a + V3b |
| 14 | M | 58 | AD | IIIA | Y | Y | EML4-ALK V3a + V3b |
| 15 | M | 62 | AD | IIIA | Y | N | EML4-ALK V3a + V3b |
| 16 | M | 38 | AD | I | Y | Y | EML4-ALK V3a + V3b |
| 17 | F | 59 | AD | I | N | N | EML4-ALK V3a + V3b |
| 18 | F | 45 | AD | I | N | Y | EML4-ALK V3a + V3b |
NSCLC, non-small cell lung cancer; AD, adenocarcinoma; ALK, anaplastic lymphoma kinase; EML4, echinoderm microtubule-associated protein-like 4; RT-PCR, reverse transcription polymerase chain reaction; M, male; F, female; Y, yes; N, no; V1, variant 1; V3a, variant 3a; V3b, variant 3b.
ROS1 rearrangement
In the 205 patients, five patients (5/205, 2.4%), one male and four females were found to harbor ROS1 rearrangement (Table 3). Most patients with ROS1 fusion were also younger, with 4 less than 55 and only 1 older than 55 years. All 5 patients had CD74-ROS1 (E6/E34) fusion gene and adenocarcinoma (Table S3), additionally none of them had co-existing ALK rearrangement. No significant association was found between ROS1 rearrangement and clinic-pathological parameters. However, all five ROS1 fusion cases experienced indoor air pollution caused by burning coal, and the one male was also a smoker.
Table S3
| No. | Gender | Age | Pathology diagnosis | Stage | Smoking history | Household coal combustion | ROS1 fusion genes detected by RT-PCR |
|---|---|---|---|---|---|---|---|
| 1 | F | 66 | AD | IIIA | N | Y | CD74-ROS1 (E6/E34) |
| 2 | F | 48 | AD | IIIA | N | Y | CD74-ROS1 (E6/E34) |
| 3 | F | 50 | AD | I | N | Y | CD74-ROS1 (E6/E34) |
| 4 | F | 51 | AD | I | N | Y | CD74-ROS1 (E6/E34) |
| 5 | M | 42 | AD | I | Y | Y | CD74-ROS1 (E6/E34) |
NSCLC, non-small cell lung cancer; RT-PCR, reverse transcription polymerase chain reaction; ROS1, ROS proto-oncogene 1; M, male; F, female; E6, exon6; E34, exon34; AD, adenocarcinoma.
The other gene rearrangement
Although potential RET rearrangement and NTRK1 rearrangement were analyzed in all subjects, no positive case was identified. Since our subject population is relatively small, with increasing sample size, it could be possible to detect other genes’ rearrangement or additional fusion partners for ALK and ROS1.
Analysis of gene fusion pattern and clinic-pathological parameters
If both ALK and ROS1 rearrangement cases were combined as gene fusion event for evaluation, total 23 cases (23/205, 11.2%) were identified, and all were adenocarcinoma (P=0.006). Most of gene fusion cases (15/23, 65.2%) happened in patients less than 55 years old (P=0.01). Moreover, gene fusion events were significantly associated with domestic coal use (P=0.004), 19 patients were coal users and only 4 patients were never coal users. When evaluated by overall exposure (tobacco and coal, union set n=145), the association was also significant (P=0.005), 22 subjects were either coal users or smokers.
Compare with other studies on EML4-ALK fusion gene
Eighteen studies which included RT-PCR for detecting ALK fusions and provided ethnicity and variant type data for EML4-ALK fusion gene were summarized (12-30) (Table S4). In the 3,535 NSCLC patients, 236 ALK rearrangement events were identified. Variant 3 (67/152, 44.1%) was the most common in Asian population, followed by variant 1 (53/152, 34.9%). Variant 1 (28/37, 75.7%) was the dominant type in Caucasian. We divided 13 Asian studies based on the gender rate and the smoking rate of ALK+ subjects (patients carrying ALK fusions) (Figure 1H), 4 quadrants represented different characters of ALK+ cases, and the bubble size indicated ALK+ rate in that study. For Asian patients, most ALK+ subjects featured with higher rate of females and nonsmokers, but the ALK+ rate in these subject populations were relatively lower (upper-left quadrant, 8 smaller bubbles). On the other hand, studies found higher male rate in ALK+ cases seemed to have increased ALK+ rate in their subject populations (upper-right quadrant, 3 larger bubbles). One Japanese study located directly on the Y axis. Present study (red bubble) set in a quadrant with few co-existing works, potentially indicating special population composition.
Table S4
| No. | Study or subgroup (reference No.) | Ethnic group | Subjects sources | Fusion gene type | Gender distribution (male/female) | Smoking history (ever/never) | Number of subjects in each variant type | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | |||||||
| 1 | Shinmura et al. 2008 (24) | Asian | Japan (n=77) | EML4-ALK (n=2) | 1/1 | 2/0 | 1 | 1 | – | – | – | – | – | – |
| 2 | Inamura et al. 2008 (17) | Asian | Japan (n=221) | EML4-ALK (n=5) | 2/3 | 2/3 | 2 | 3 | – | – | – | – | – | – |
| 3 | Koivunen et al. 2008 (19) | Asian | Korea (n=167) | EML4-ALK (n=8) | 3/5 | 2/6 | 2 | – | 4 | 2 | – | – | – | – |
| 4 | Takeuchi et al. 2008 (26)a; Inamura et al. 2009 (16)a |
Asian | Japan (n=363) | EML4-ALK (n=11) | 5/6 | 1/10 | 3 | 3 | 3 | 1 | 1 | – | – | – |
| 5 | Takeuchi et al. 2009 (27) | Asian | Japan (n=130) | EML4-ALK (n=7); KIF5B-ALK (n=1) | NA | NA | 1 | 1 | 3 | – | – | 1 | 1 | – |
| 6 | Wong et al. 2009 (29) | Asian | China (n=266) | EML4-ALK (n=13) | 5/8 | 1/12 | 2 | 2 | 8 | – | 1 | – | – | – |
| 7 | Takahashi et al. 2010 (25) | Asian | Japan (n=313) | EML4-ALK (n=5)b | 1/4 | 1/4 | 1 | – | 2 | – | – | – | – | – |
| 8 | Zhang et al. 2010 (30) | Asian | China (n=103) | EML4-ALK (n=12) | 7/5 | 2/10 | 4 | 1 | 3 | – | 1 | 3 | – | – |
| 9 | Jin et al. 2012 (18) | Asian | Korea (n=167) | EML4-ALK (n=10) | 4/6 | 3/7 | 8 | – | 2 | – | – | – | – | – |
| 10 | Li et al. 2013 (20) | Asian | China (n=208) | EML4-ALK (n=7) | 0/7 | 1/6 | 2 | 1 | 4 | – | – | – | – | – |
| 11 | Wu et al. 2013 (14) | Asian | China (n=312) | EML4-ALK (n=12); KIF5B-ALK (n=1) | 4/8 | 5/7 | 3 | 1 | 8 | – | – | – | – | – |
| 12 | Wang et al. 2014(28) | Asian | China(n=430) | EML4-ALK (n=62)c; KIF5B-ALK (n=2) | 25/21 | 17/29 | 24 | 8 | 30 | – | – | – | – | – |
| 13 | Shan et al. 2014 (12) | Asian | China(n=297) | EML4-ALK (n=37)d | 20/17 | 9/22 (6: NA) | – | – | – | – | – | – | – | – |
| Total for Asian | – | 3,054 | 195 | 77/91 | 46/116 | 53 | 21 | 67 | 3 | 3 | 4 | 1 | – | |
| 14 | Lin et al. 2009 (21) | Caucasian | USA (n=106) | EML4-ALK (n=12) | NA | NA | 11 | – | – | – | – | – | – | 1 |
| 15 | Martelli et al. 2009 (22) | Caucasian | Italy and Spain (n=120) | EML4-ALK (n=9) | 8/1 | 8/1 | 7 | – | 2 | – | – | – | – | – |
| 16 | Sanders et al. 2011 (23) | Caucasian | USA (n=55) | EML4-ALK (n=5) | NA | NA | 1 | – | 3 | – | – | – | – | 1 |
| 17 | Hofman et al. 2012 (15) | Caucasian | France (n=154) | EML4-ALK (n=4)d | 0/4 | 0/4 | – | – | – | – | – | – | – | – |
| 18 | Wallander et al. 2012 (13) | Caucasian | USA (n=46) | EML4-ALK (n=11)e | 11/5 | NA | 9 | – | 2 | – | – | – | – | – |
a, there were overlapping in the subject sources, so the data were shown as combined together; b, two of the 5 subjects positive for EML4-ALK had no variant type data (the authors considered them to be novel types); c, EML4-ALK variant types were detected by RT-PCR, characteristics of patients were reported based on FISH results; d, the study did not give the data of EML4-ALK variant types; e, 11 subjects positive for EML4-ALK had variant type data, the other 5 was identified by FISH. EML4, echinoderm microtubule-associated protein-like 4; ALK, anaplastic lymphoma kinase; KIF5B, kinesin family member 5B; RT-PCR, reverse transcription polymerase chain reaction; FISH: fluorescence in situ hybridization; NA: Data not included in the report; V1, variant 1; V2, variant 2; V3, variant 3; V4, variant 4; V5, variant 5; V6, variant 6; V7, variant 7; V8, variant 8.
Discussion
ALK signal pathway involves cell proliferation, differentiation, and anti-apoptosis (11,14). Various ALK rearrangements have been discovered in different neoplasms, the EML4-ALK fusion gene is recognized as an important oncogenic driver in NSCLC (11,19,26,28). The incidence of EML4-ALK fusion in NSCLC was around 0.99% to 15%, with no significant differences between Asian and western countries (12-16,18,24-36). Most of our results were consistent with previous reports: 8.78% ALK rearrangement in NSCLC patients, ALK+ cases tended to be younger and variant 3 was the dominant type. In Asian populations, EML4-ALK fusion variant 3 had an average rate of 44% (range, 20–67%), but our study found a much higher variant 3 frequency (77.8%). After further comparison with previous references for gender, smoking and ALK+ rate, present study fell into an independent quadrant with few co-existing report (Figure 1H). It might reflect the unique features of our subject population, since most ALK+ patients were coal users.
Many previous Asian reports (14,22,24,28) found ALK rearrangement had association with female gender and nonsmoker, but some showed balance between both genders (12,18,25,29). One study including 1,178 ALK rearranged cases from 20,541 NSCLC patients (11) indicated: gender might be potential source of study heterogeneity. Our study indicated ALK rearrangement was significantly associated with HCU (P=0.048) and overall exposure (P=0.027). There could be similar mechanism behind our finding and others: in Asian most females are nonsmokers, but in rural areas females are usually responsible for cooking, thus exposed more frequently and severely to indoor air pollution from coal use. Coal-burning release a cocktail of carcinogens, like polycyclic aromatic hydrocarbons (PAHs), fine particulate matter (PM2.5), many of which are class I carcinogens defined by International Agency for Research on Cancer (IARC) (4,6,31). On the other hand, the number of carcinogens released by cigarette may not be comparable to coal-burning, especially when burned indoor. As a result, there could possibly be more gene fusion events in females. The assumption at least may partly explain EML4-ALK fusion gene was associated with female gender in some studies.
ROS1 rearrangement characterizes a small subset (range, 0.5–2%) of NSCLC, and is associated with slight or nonsmokers and adenocarcinoma. Even ROS1 can have different fusion partners, but CD74-ROS1 is the major type (~40%), follow by EZR-ROS1 and unknown type (both around ~15%) (32). What we found about ROS1 fusions were quite similar to previous reports (32-34), except that our ROS1 fusions were all CD74-ROS1 (E6/E34) and happened in subjects affected by indoor coal-using. Whether this could be a feature of our subject population still require further study.
When all the gene rearrangement events were combined for evaluation, it showed association with HCU and overall exposure level (tobacco and coal), suggesting coal-burning could possibly be a vital player in causing gene rearrangement. Similarly, other researches on lung cancer conducted in this region found higher mutation rate in KRAS (range, 15–29%) (7,9), but KRAS mutations are lower in other Asian populations (range, 2–7%), including smokers and nonsmokers (35-38). Coal combustion releases a variety of carcinogens, each may have different effect, for example, p53 mutations from nonsmoking females in Xuanwei were consistent with those induced by PAHs, and different from those attributes to smoking (9). Current study and other reports (2,7,35,37) all support there could be unique mutational patterns in lung cancer subjects exposed to coal combustion.
Conclusions
Present work found clinic-pathologic features and gene fusion patterns of ALK and ROS1 in NSCLC show association with HCU. Our findings may help evaluate the impact of coal combustion on the pathogenesis of lung cancer, and also highlight the importance of integrating various environmental/occupational factors into clinical and theoretical research.
Supplementary
PCR conditions for each fusion gene
ALK rearrangement
Two pairs of primers (Table S1) were designed based on references to identify different EML4-ALK Variants (14-16). The first was to detect variant 1, and the second was to cover EML4-ALK variant 2 to 7. The thermal cycle conditions were also different. For EML4-ALK Variant 1, the program was: 5 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 55 °C, 60 seconds at 72 °C. For EML4-ALK Variant 2–7, the program was: 10 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 66 °C, 2.5 minute at 72 °C.
Table S1
| Gene and variant | Sequence |
|---|---|
| EML4-ALK, Variant 1 | Forward: 5'-GTGCAGTGTTTAGCATTCTTGGGG-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| EML4-ALK, Variant 2–7 | Forward: 5'-GTCAGCTCTTGAGTCACGAGTT-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| KIF5B-ALK | Forward: 5'-TCGGCAACTTTAGCGAGTA-3' |
| Reverse: 5'-GGACACCTGGCCTTCATAC-3' | |
| KLC1-ALK | Forward: 5'-ATGTATGACAACATGTCCAC-3' |
| Reverse: 5'-TCAGGGCCCAGGCTGGTTCA-3' | |
| TFG-ALK | Forward: 5'-TCGTTTATTGGATAGCTTGGAACCAC-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| TPM3-ROS1 (E10/E35) | Forward: 5'-CTTACTGATAAACTCAAGGAGGCAGAGAC-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| SDC4-ROS1 (E2/E32) | Forward: 5'-CAGGACCTCCTAGAAGGCCGATAC-3' |
| Reverse: 5'-CATCCATTATCTTCAGCTTTCTCCCACT-3' | |
| SDC4-ROS1 (E4/E32) | Forward: 5'-ACCCGTTGAAGAGAGTGAGGATG-3' |
| Reverse: 5'-TGAAGTGCTCTTTCTTATCTCAAGG-3' | |
| SDC4-ROS1 (E4/E34) | Forward: 5'-AGGATGTGTCCAACAAGGTGTCAATG-3' |
| Reverse: 5'-TAATCTTCTATGCCAGACAAAGGTCAGTG-3' | |
| SLC34A2-ROS1 (E13/E32) | Forward: 5'-GTGCTGCTTGCTGTGTGGCT-3' |
| Reverse: 5'-CTACATCCATTATCTTCAGCTTTCTCCCA-3' | |
| CD74-ROS1 (E6/E32) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CATTATCTTCAGCTTTCTCCCACTGTATTG-3' | |
| CD74-ROS1 (E6/E34) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CTCTTTGTCTTCGTTTATAAGCACTGTCAC-3' | |
| EZR-ROS1 (E10/E34) | Forward: 5'-AAAGGAGAGAAACCGTGGAGAGAG-3' |
| Reverse: 5'-GCCAGACAAAGGTCAGTGGGATT-3' | |
| LRIG3-ROS1 (E17/E35) | Forward: 5'-GACCAACTTGCCAGCAGATATTCCT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| GOPC-ROS1 (E7:E35) | Forward: 5'-CGTAGAGTATGAAGATGAGAGTGGACAT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| CCDC6-RET | Forward: 5'-GCTGGAGACCTACAAACTGA-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| KIF5B-RET | Forward: 5'-TAAGGAAATGACCAACCACCAG-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| CD74-NTRK1 | Forward: 5'-CTCCCAAGCCTGTGAGCAAGAT-3' |
| Reverse: 5'-GTTGTGGCACTCAGCAAGGAAG-3' |
The potential existence of (TRK-fused gene) TFG-ALK, (kinesin light chain 1) KLC1-ALK and (kinesin family member 5B) KIF5B-ALK, were also examined in all samples that were negative for EML4-ALK fusion gene. The primers (Table S1) and PCR conditions were designed according to previous reports (17-19). The program for KIF5B-ALK was: 5 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 50 °C, and 2.5 minutes at 72 °C. For KLC1-ALK, the program was 5 minutes at 95 °C, followed by 40 cycles of 40 seconds at 95 °C, 40 seconds at 55 °C and 3 minutes at 72 °C. For TFG-ALK, it was 5 minutes at 95 °C, followed by 40 cycles of 40 seconds at 94 °C, 40 seconds at 65 °C, and 1.5 minutes at 72 °C.
ROS1 rearrangement
In order to detect potential ROS1 fusions, including CD74-ROS1, TPM3-ROS1 (tropomyosin 3), EZR-ROS1 (ezrin), SLC34A2-ROS1 (solute carrier family 34 member 2), LRIG3-ROS1 (leucine rich repeats and immunoglobulin like domains 3), GOPC-ROS1 (golgi associated PDZ and coiled-coil motif containing), and SDC4-ROS1 (syndecan 4). The primers (Table S1) and PCR condition were designed based on previous studies (20,21). PCR condition is: 5 min at 95 °C, followed by 10 cycles of touchdown PCR (annealing temperature from 63 °C to 58 °C with a 0.5 decrease each cycle) and 30 cycles of 40 seconds at 95 °C, 40 seconds at 58 °C, 1 minute at 72 °C, with a final extension of 72 °C for 5 min.
RET rearrangement
The potential existence of CCDC6-RET (coiled-coil domain containing 6) and KIF5B-RET, were also examined in all samples. The primers (Table S1) and PCR conditions were designed according to previous study (22). The program to detect CCDC6-RET was: 4 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 60 °C, 30 seconds at 72 °C. The program for KIF5B-RET fusion was: 4 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 62 °C, 30 seconds at 72 °C.
NTRK1 rearrangement
In an attempt to detect the rarely reported CD74-NTRK1 fusion, the primers (Table S1) and PCR condition were set based on the previous report (23). PCR condition was: 4 min at 95 °C, followed by 10 cycles of touchdown PCR (annealing temperature from 62 °C to 57 °C with a 0.5 decrease each cycle) and 30 cycles of 40 seconds at 95 °C, 40 seconds at 57 °C, 1 minute at 72 °C.
After PCR reactions, PCR products were first examined by electrophoresis, any PCR product with expected size was selected out for Sanger sequencing confirmation by BGI Tech, Shenzhen, China (http://www.bgitechsolutions.com). Sequences obtained were verified via NCBI-BLAST. All positive fusion cases were confirmed by another independent PCR reaction.
ALK rearrangement
Two pairs of primers (Table S1) were designed based on references to identify different EML4-ALK Variants (14-16). The first was to detect variant 1, and the second was to cover EML4-ALK variant 2 to 7. The thermal cycle conditions were also different. For EML4-ALK Variant 1, the program was: 5 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 55 °C, 60 seconds at 72 °C. For EML4-ALK Variant 2–7, the program was: 10 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 66 °C, 2.5 minute at 72 °C.
Table S1
| Gene and variant | Sequence |
|---|---|
| EML4-ALK, Variant 1 | Forward: 5'-GTGCAGTGTTTAGCATTCTTGGGG-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| EML4-ALK, Variant 2–7 | Forward: 5'-GTCAGCTCTTGAGTCACGAGTT-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| KIF5B-ALK | Forward: 5'-TCGGCAACTTTAGCGAGTA-3' |
| Reverse: 5'-GGACACCTGGCCTTCATAC-3' | |
| KLC1-ALK | Forward: 5'-ATGTATGACAACATGTCCAC-3' |
| Reverse: 5'-TCAGGGCCCAGGCTGGTTCA-3' | |
| TFG-ALK | Forward: 5'-TCGTTTATTGGATAGCTTGGAACCAC-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| TPM3-ROS1 (E10/E35) | Forward: 5'-CTTACTGATAAACTCAAGGAGGCAGAGAC-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| SDC4-ROS1 (E2/E32) | Forward: 5'-CAGGACCTCCTAGAAGGCCGATAC-3' |
| Reverse: 5'-CATCCATTATCTTCAGCTTTCTCCCACT-3' | |
| SDC4-ROS1 (E4/E32) | Forward: 5'-ACCCGTTGAAGAGAGTGAGGATG-3' |
| Reverse: 5'-TGAAGTGCTCTTTCTTATCTCAAGG-3' | |
| SDC4-ROS1 (E4/E34) | Forward: 5'-AGGATGTGTCCAACAAGGTGTCAATG-3' |
| Reverse: 5'-TAATCTTCTATGCCAGACAAAGGTCAGTG-3' | |
| SLC34A2-ROS1 (E13/E32) | Forward: 5'-GTGCTGCTTGCTGTGTGGCT-3' |
| Reverse: 5'-CTACATCCATTATCTTCAGCTTTCTCCCA-3' | |
| CD74-ROS1 (E6/E32) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CATTATCTTCAGCTTTCTCCCACTGTATTG-3' | |
| CD74-ROS1 (E6/E34) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CTCTTTGTCTTCGTTTATAAGCACTGTCAC-3' | |
| EZR-ROS1 (E10/E34) | Forward: 5'-AAAGGAGAGAAACCGTGGAGAGAG-3' |
| Reverse: 5'-GCCAGACAAAGGTCAGTGGGATT-3' | |
| LRIG3-ROS1 (E17/E35) | Forward: 5'-GACCAACTTGCCAGCAGATATTCCT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| GOPC-ROS1 (E7:E35) | Forward: 5'-CGTAGAGTATGAAGATGAGAGTGGACAT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| CCDC6-RET | Forward: 5'-GCTGGAGACCTACAAACTGA-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| KIF5B-RET | Forward: 5'-TAAGGAAATGACCAACCACCAG-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| CD74-NTRK1 | Forward: 5'-CTCCCAAGCCTGTGAGCAAGAT-3' |
| Reverse: 5'-GTTGTGGCACTCAGCAAGGAAG-3' |
The potential existence of (TRK-fused gene) TFG-ALK, (kinesin light chain 1) KLC1-ALK and (kinesin family member 5B) KIF5B-ALK, were also examined in all samples that were negative for EML4-ALK fusion gene. The primers (Table S1) and PCR conditions were designed according to previous reports (17-19). The program for KIF5B-ALK was: 5 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 50 °C, and 2.5 minutes at 72 °C. For KLC1-ALK, the program was 5 minutes at 95 °C, followed by 40 cycles of 40 seconds at 95 °C, 40 seconds at 55 °C and 3 minutes at 72 °C. For TFG-ALK, it was 5 minutes at 95 °C, followed by 40 cycles of 40 seconds at 94 °C, 40 seconds at 65 °C, and 1.5 minutes at 72 °C.
ROS1 rearrangement
In order to detect potential ROS1 fusions, including CD74-ROS1, TPM3-ROS1 (tropomyosin 3), EZR-ROS1 (ezrin), SLC34A2-ROS1 (solute carrier family 34 member 2), LRIG3-ROS1 (leucine rich repeats and immunoglobulin like domains 3), GOPC-ROS1 (golgi associated PDZ and coiled-coil motif containing), and SDC4-ROS1 (syndecan 4). The primers (Table S1) and PCR condition were designed based on previous studies (20,21). PCR condition is: 5 min at 95 °C, followed by 10 cycles of touchdown PCR (annealing temperature from 63 °C to 58 °C with a 0.5 decrease each cycle) and 30 cycles of 40 seconds at 95 °C, 40 seconds at 58 °C, 1 minute at 72 °C, with a final extension of 72 °C for 5 min.
RET rearrangement
The potential existence of CCDC6-RET (coiled-coil domain containing 6) and KIF5B-RET, were also examined in all samples. The primers (Table S1) and PCR conditions were designed according to previous study (22). The program to detect CCDC6-RET was: 4 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 60 °C, 30 seconds at 72 °C. The program for KIF5B-RET fusion was: 4 minutes at 95 °C, followed by 40 cycles of 30 seconds at 95 °C, 30 seconds at 62 °C, 30 seconds at 72 °C.
NTRK1 rearrangement
In an attempt to detect the rarely reported CD74-NTRK1 fusion, the primers (Table S1) and PCR condition were set based on the previous report (23). PCR condition was: 4 min at 95 °C, followed by 10 cycles of touchdown PCR (annealing temperature from 62 °C to 57 °C with a 0.5 decrease each cycle) and 30 cycles of 40 seconds at 95 °C, 40 seconds at 57 °C, 1 minute at 72 °C.
After PCR reactions, PCR products were first examined by electrophoresis, any PCR product with expected size was selected out for Sanger sequencing confirmation by BGI Tech, Shenzhen, China (http://www.bgitechsolutions.com). Sequences obtained were verified via NCBI-BLAST. All positive fusion cases were confirmed by another independent PCR reaction.
Supplementary
Table S1
| Gene and variant | Sequence |
|---|---|
| EML4-ALK, Variant 1 | Forward: 5'-GTGCAGTGTTTAGCATTCTTGGGG-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| EML4-ALK, Variant 2–7 | Forward: 5'-GTCAGCTCTTGAGTCACGAGTT-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| KIF5B-ALK | Forward: 5'-TCGGCAACTTTAGCGAGTA-3' |
| Reverse: 5'-GGACACCTGGCCTTCATAC-3' | |
| KLC1-ALK | Forward: 5'-ATGTATGACAACATGTCCAC-3' |
| Reverse: 5'-TCAGGGCCCAGGCTGGTTCA-3' | |
| TFG-ALK | Forward: 5'-TCGTTTATTGGATAGCTTGGAACCAC-3' |
| Reverse: 5'-TCTTGCCAGCAAAGCAGTAGTTGG-3' | |
| TPM3-ROS1 (E10/E35) | Forward: 5'-CTTACTGATAAACTCAAGGAGGCAGAGAC-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| SDC4-ROS1 (E2/E32) | Forward: 5'-CAGGACCTCCTAGAAGGCCGATAC-3' |
| Reverse: 5'-CATCCATTATCTTCAGCTTTCTCCCACT-3' | |
| SDC4-ROS1 (E4/E32) | Forward: 5'-ACCCGTTGAAGAGAGTGAGGATG-3' |
| Reverse: 5'-TGAAGTGCTCTTTCTTATCTCAAGG-3' | |
| SDC4-ROS1 (E4/E34) | Forward: 5'-AGGATGTGTCCAACAAGGTGTCAATG-3' |
| Reverse: 5'-TAATCTTCTATGCCAGACAAAGGTCAGTG-3' | |
| SLC34A2-ROS1 (E13/E32) | Forward: 5'-GTGCTGCTTGCTGTGTGGCT-3' |
| Reverse: 5'-CTACATCCATTATCTTCAGCTTTCTCCCA-3' | |
| CD74-ROS1 (E6/E32) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CATTATCTTCAGCTTTCTCCCACTGTATTG-3' | |
| CD74-ROS1 (E6/E34) | Forward: 5'-GGATGCACCATTGGCTCCTGTT-3' |
| Reverse: 5'-CTCTTTGTCTTCGTTTATAAGCACTGTCAC-3' | |
| EZR-ROS1 (E10/E34) | Forward: 5'-AAAGGAGAGAAACCGTGGAGAGAG-3' |
| Reverse: 5'-GCCAGACAAAGGTCAGTGGGATT-3' | |
| LRIG3-ROS1 (E17/E35) | Forward: 5'-GACCAACTTGCCAGCAGATATTCCT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| GOPC-ROS1 (E7:E35) | Forward: 5'-CGTAGAGTATGAAGATGAGAGTGGACAT-3' |
| Reverse: 5'-ACCTCGCAGCTCAGCCAACT-3' | |
| CCDC6-RET | Forward: 5'-GCTGGAGACCTACAAACTGA-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| KIF5B-RET | Forward: 5'-TAAGGAAATGACCAACCACCAG-3' |
| Reverse: 5'-CCTTGACCACTTTTCCAAATTC-3' | |
| CD74-NTRK1 | Forward: 5'-CTCCCAAGCCTGTGAGCAAGAT-3' |
| Reverse: 5'-GTTGTGGCACTCAGCAAGGAAG-3' |
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.37). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ethical committees of Yunnan Cancer Hospital approved the study (No.KY2019.57). All patients provided informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Ezzati M, Kammen DM. The health impacts of exposure to indoor air pollution from solid fuels in developing countries: knowledge, gaps, and data needs. Environ Health Perspect 2002;110:1057-68. [Crossref] [PubMed]
- Zhang JJ, Smith KR. Household air pollution from coal and biomass fuels in China: measurements, health impacts, and interventions. Environ Health Perspect 2007;115:848-55. [Crossref] [PubMed]
- Hosgood HD, Boffetta P, Greenland S, et al. In-home coal and wood use and lung cancer risk: a pooled analysis of the International Lung Cancer Consortium. Environ Health Perspect 2010;118:1743-7. [Crossref] [PubMed]
- Mumford JL, He XZ, Chapman RS, et al. Lung cancer and indoor air pollution in Xuan Wei, China. Science 1987;235:217-20. [Crossref] [PubMed]
- Hosgood HD, Pao W, Rothman N, et al. Driver mutations among never smoking female lung cancer tissues in China identify unique EGFR and KRAS mutation pattern associated with household coal burning. Respir Med 2013;107:1755-62. [Crossref] [PubMed]
- Lee YJ, Kim JH, Kim SK, et al. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer 2011;72:9-15. [Crossref] [PubMed]
- DeMarini DM, Landi S, Tian D, et al. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res 2001;61:6679-81. [PubMed]
- Wang TW, Vermeulen RCH, Hu W, et al. Gene-expression profiling of buccal epithelium among non-smoking women exposed to household air pollution from smoky coal. Carcinogenesis 2015;36:1494-501. [PubMed]
- Fan L, Feng Y, Wan H, et al. Clinicopathological and demographical characteristics of non-small cell lung cancer patients with ALK rearrangements: a systematic review and meta-analysis. PLoS One 2014;9:e100866. [Crossref] [PubMed]
- Shan L, Lian F, Guo L, et al. Combination of conventional immunohistochemistry and qRT-PCR to detect ALK rearrangement. Diagn Pathol 2014;9:3. [Crossref] [PubMed]
- Wallander ML, Geiersbach KB, Tripp SR, et al. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med 2012;136:796-803. [Crossref] [PubMed]
- Wu YC, Chang IC, Wang CL, et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS One 2013;8:e70839. [Crossref] [PubMed]
- Hofman P, Ilie M, Hofman V, et al. Immunohistochemistry to identify EGFR mutations or ALK rearrangements in patients with lung adenocarcinoma. Ann Oncol 2012;23:1738-43. [Crossref] [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009;22:508-15. [Crossref] [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008;3:13-7. [Crossref] [PubMed]
- Jin G, Jeon HS, Lee EB, et al. EML4-ALK fusion gene in Korean non-small cell lung cancer. J Korean Med Sci 2012;27:228-30. [Crossref] [PubMed]
- Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008;14:4275-83. [Crossref] [PubMed]
- Li Y, Li Y, Yang T, et al. Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One 2013;8:e52093. [Crossref] [PubMed]
- Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res 2009;7:1466-76. [Crossref] [PubMed]
- Martelli MP, Sozzi G, Hernandez L, et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol 2009;174:661-70. [Crossref] [PubMed]
- Sanders HR, Li HR, Bruey JM, et al. Exon scanning by reverse transcriptase-polymerase chain reaction for detection of known and novel EML4-ALK fusion variants in non-small cell lung cancer. Cancer Genet 2011;204:45-52. [Crossref] [PubMed]
- Shinmura K, Kageyama S, Tao H, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 2008;61:163-9. [Crossref] [PubMed]
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 2010;17:889-97. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618-24. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143-9. [Crossref] [PubMed]
- Wang J, Cai Y, Dong Y, et al. Clinical characteristics and outcomes of patients with primary lung adenocarcinoma harboring ALK rearrangements detected by FISH, IHC, and RT-PCR. PLoS One 2014;9:e101551. [Crossref] [PubMed]
- Wong DWS, Leung ELH, So KKT, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 2009;115:1723-33. [Crossref] [PubMed]
- Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 2010;9:188. [Crossref] [PubMed]
- Gordon SB, Bruce NG, Grigg J, et al. Respiratory risks from household air pollution in low and middle income countries. Lancet Respir Med 2014;2:823-60. [Crossref] [PubMed]
- Rossi G, Jocollé G, Conti A, et al. Detection of ROS1 rearrangement in non-small cell lung cancer: current and future perspectives. Lung Cancer (Auckl) 2017;8:45-55. [Crossref] [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [Crossref] [PubMed]
- Fu S, Liang Y, Lin YB, et al. The Frequency and Clinical Implication of ROS1 and RET Rearrangements in Resected Stage IIIA-N2 Non-Small Cell Lung Cancer Patients. PLoS One 2015;10:e0124354. [Crossref] [PubMed]
- Kawaguchi T, Ando M, Kubo A, et al. Long exposure of environmental tobacco smoke associated with activating EGFR mutations in never-smokers with non-small cell lung cancer. Clin Cancer Res 2011;17:39-45. [Crossref] [PubMed]
- Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer 2008;99:245-52. [Crossref] [PubMed]
- Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. [Crossref] [PubMed]
- Wu CC, Hsu HY, Liu HP, et al. Reversed mutation rates of KRAS and EGFR genes in adenocarcinoma of the lung in Taiwan and their implications. Cancer 2008;113:3199-208. [Crossref] [PubMed]


