
The regulatory role of aberrant methylation of microRNA-34a promoter CpGs in osteosarcoma
Introduction
As the most common bone-derived malignancy of adolescents, osteosarcoma is highly invasive and metastatic. Because the exact pathogenesis of osteosarcoma is unknown and the rate of early diagnosis is low, the disease easily relapses and metastasizes. The 5-year survival rate of osteosarcoma is only 60% to 70% (1). It is known that osteosarcoma is caused by genetic mutations and malignant proliferation of mesenchymal cells with osteogenic potential, and genetic and epigenetic changes are involved in this process. Therefore, the discovery of key molecular biologic markers in the development of osteosarcoma and early identification and effective target blockade of these markers are extremely important for early diagnosis and effective treatment.
The role of microRNAs (miRNAs) in malignant tumors has become increasingly significant because they participate in cell growth, development, apoptosis, and other complex life processes (2). Although the specific mechanisms of miRNAs in carcinogenesis should be further investigated, it is clear that this process involves the inhibition of target gene expression, induction of messenger RNA (mRNA) degradation by complementing with target genes, and the influence of the promoter CpGs methylation participating in the transcriptional regulation process (3). MiR-34 plays an important role in the induction of apoptosis, cell cycle arrest, and cell senescence, for example, acting as direct targets of p53 (4-6). As a member of the miR-34s family, miR-34a acts as a tumor suppressor by negatively regulating a variety of important regulatory proteins, such as E2F3, c-MYC, Bcl2, c-MET, and CDK4/6 (7,8). The results of in-vitro and in-vivo studies have also proved that miR-34a can inhibit the growth and metastasis of osteosarcoma cells by regulating c-Met, Eag1, and other molecules in osteosarcoma cells (9,10).
DNA methylation is one of the important modification pathways in epigenetics, which influences biological processes such as the structure of chromatin, the stability of DNA, and the interaction between DNA and protein (11). About 60% of the gene promoter regions contain CpGs, but only a portion of the CpGs5'-end cytosines are converted to 5-methylcytosine. These methylated CpGs cause chromatin inactivation by histone deacetylase, and thus maintain biologically specific expression (12). However, during oncogenesis, the abnormal methylation of CpGs occurs in some promoter regions of the key tumor suppressor gene, which is stably inherited into the next generation of cells for selective cell clone amplification to cause tumor cell growth (2,13,14). Some reports showed that abnormal CpGs methylation occurs during the early stages of tumor progression, so its detection may be used in the future for the evaluation of tumor diagnosis and prognosis (15).
The DNA methylation process is catalyzed by several DNA methyltransferases, in which DNMT1 is responsible for accurately replicating and maintaining the pre-existing methylation pattern following DNA replication. In the study of cervical cancer, DNMT1 could inhibit the transcription of tumor suppressor genes and promote tumor formation (16). Therefore, it can be speculated that the inhibition of DNA methyltransferase activity may reduce hypermethylation of the suppressor gene promoter and thus may promote its re-expression to reverse the phenotype of malignant tumors. Studies on colorectal, kidney, pancreatic, breast, and uterine cancers, and soft tissue sarcoma have shown that miR-34a expression of the tumor tissue was lower than that of normal tissue, and it was accompanied with abnormal methylation of the miR-34a promoter CpGs (12). However, the process of methylation of the miR-34a promoter CpGs in osteosarcoma and its regulatory role are not yet clear. The gene mutation in the normal human body is an important cause of tumorigenesis, while results on gene detection of tumor tissue and its peripheral tissue are not quite persuasive. Therefore, we designed a study to investigate the difference between osteosarcoma and normal human bone tissues in the methylation of miR-34a promoter CpGs, and further study the regulatory mechanism of miR-34a and its promoter methylation in osteosarcoma.
Methods
Tissue specimens
Tissue specimens were collected from patients with osteosarcoma who were admitted to the General Hospital of Ningxia Medical University, Yinchuan City, Ningxia Province, China, from June 2010 to June 2016. Osteosarcoma tissue specimens were collected from the distal femur or proximal tibia of patients who were diagnosed with osteosarcoma, based on the pathological examination after tumor resection. No patient had received radiotherapy or chemotherapy before surgery. Normal bone tissue specimens were collected from the distal femur or proximal tibia of 12 patients who underwent artificial knee arthroplasty. All tissue specimens were snap-frozen in liquid nitrogen after surgical resection.
This study was approved by the Ethics Committee of the General Hospital of Ningxia Medical University. All tissue specimens were collected after informed consent was obtained from each subject, and all specimens were treated according to legal standards for ethical handling and patient anonymity.
Cell culture
The osteosarcoma cell lines MG-63 and SAOS-2 were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). ATTC MG-63 was cultured using Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Shelton, CT, USA), and SAOS-2 was cultured using McCoy’s 5A Medium (Gibco, USA). The osteoblast cell line hFOB1.19 was also purchased from ATCC and cultured using DMEM medium (Gibco, USA). All culture media were supplemented with 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin and 100 g/mL streptomycin, and cultured in a 5% CO2 incubator at 37 °C.
Reagents and transfection
An epigenetic modifier, 5-aza-2'-deoxycytidine (5-aza-dC) and the MTT kit were purchased from Sigma-Aldrich (USA). Two chemically synthesized double-stranded RNA oligonucleotides (miR-34a mimics and mimics NC) were purchased from Ribo Biotech, Ltd (Guangzhou, China). MiRNA mimics can simulate high-level expression of mature miRNA in cells to enhance the regulation of endogenous miRNAs. Transfection reagent LipofectamineTM 2000 was purchased from Ribo Biotech, Ltd. (China). The cell count before transfection was 3.0×105 cells/35 mm petri dish. The small RNA molecules labeled with Cy3 dye were diluted with 200 µL of Opti-MEM (Gibco), and then 2 µL of the transfection reagent was well mixed with 200 µL Opti-MEM. The mixture was let stand for 3 to 5 minutes, and the two substances were well mixed and then let stand for 15 minutes, to make a 400 µL mixture (the Cy3-labeled small-molecule RNA mother liquor was 20 µM; 2 µL was used, and the final transfection concentration was 50 nM). The culture medium in the 35-mm Petri dish was discarded, 400 µL of fresh medium was added, and then 400 µL of the above-mentioned mixture was added, to make up 800 µL of mixture. After 24 hours of transfection, it was washed with lead sulfide (PbS), added with 1,500 µL fresh medium, and observed under a microscope.
DNA extraction and bisulfite treatment
The genomic DNA of cells and tissue specimens was isolated using Protease K and a QIAamp DNA Mini Kit (Qiagen Inc, Valencia, CA, USA) according to the manufacturer’s instructions. All purified genomic DNA specimens were quantified by spectrophotometry. 100 ng of DNA was obtained and the quality inspection was performed using 0.8% agarose gel electrophoresis. The electrophoretic bands of the genomic DNA were more than 20 kilobytes (kb), A260/A280=1.7–2.1, and the total amount was more than 2 µg, which indicated that the main band of the electrophoresis was not degraded and could be used for detecting the distribution of the miR-34a promoter methylation. The genomic DNA concentration was adjusted to 75 ng/µL, and the specimen was transferred to a 384-well plate and stored at –20 °C. The bisulfite modification of the genomic DNA was carried out using the EZ DNA Methylation-Gold™ Kit (Zymo Research Corp, Irvine, CA, USA), according to the manufacturer’s instructions, which combined the bisulfite conversion and DNA cleavage.
Quantitative analysis of DNA methylation
The quantitative analysis of miR-34a promoter CpGs methylation was performed using the Sequenom MassARRAY iPLEX™ platform (CapitalBio, Beijing, China). Primers were designed by the Sequenom EpiDesigner (http://www.epidesigner.com) and synthesized by a biological company (ANNEX 1). The 10mer tag was added to the forward 5' end of the designed primers to equilibrate the PCR conditions. The T7-Promoter sequence was added to the reverse 5' end for in-vitro transcription (IVT). The 5'-primer and 3'-(T7) primer were mixed to a final concentration of 1 µmol/L. Polymerase chain reaction (PCR) amplification was performed after bisulfite DNA modification using the PCR Accessory Set (Sequenom, 11327), according to the manufacturer’s instructions. At the end of PCR amplification, electrophoresis of the PCR product was performed using 2% agarose gel. The 5 µL 2% agarose gel was placed on the electrophoresis plate with 2× Loading Buffer, and then 1 µL PCR products was added. The result of amplification was identified after electrophoresis. Alkaline phosphatase treatment (SAP), IVT, and RNase digestion were performed according to the instructions using the MassCLEAVE Kit (Sequenom, 10129.1). The digested products were purified using the SpectroCHIP® Arrays and Clean Resin Kit (Sequenom, 10117) according to the manufacturer’s instructions, and then spotted onto a 384-element chip (SpectroCHIP, Sequenom) using the MassARRAY Nanodispenser RS1000 spotter (Sequenom), the prepared chip was placed into the MassARRAY Compact System (Sequenom) for testing. The MALDI-TOF technique was used for chip analysis, and the obtained data were treated with the Epityper software version 1.0 (Sequenom, San Diego, CA, USA).
RNA extraction and real-time PCR
RNA was extracted from cells and tissue specimens using TRIzol® (Invitrogen®, Thermo-Fisher Scientific) according to the manufacturer’s instructions. cDNA reverse transcription and real-time reverse transcription-polymerase chain reaction (RT-PCR) were performed with the miRNA qPCR Starter Kit (Ribo Biotech) and iQ™SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) on a Bio-Rad CFX 96™ real-time assay system using the RNA extracted from the specimens as a template. The small nuclear RNA U6 gene was taken as an internal control. The relative expression of miRNAs was calculated as 2−[(Ct ofmiR-34a) – (Ct ofU6)] based on Ct after normal expression of U6. The miRNA quantification was carried out with the Bulge-Loop™ miRNA qRT-PCR Primer Set, miR-34a-specific primer groups (each group had a RT primer and a pair of qPCR primers) designed by Ribo-Biotech, Ltd., Guangzhou, Guangdong, China.
Statistical analysis
The study contained at least three independent comparative experiments, and all data were represented as mean ± standard deviation (SD). SPSS Statistics 17.0 software was used for data analysis. Paired t-test was used for comparison of two groups, and one-way analysis of variance (ANOVA) was used for the comparison of multiple groups. A significance level of 0.05 was adopted.
Results
Some miR-34a promoter CpGs in osteosarcoma were abnormally hypermethylated and associated with abnormal expression of miR-34a
The MassARRAY system can be used as a tool for high-throughput detection and quantitative analysis of methylation of a single CpG at the target fragment, which can generate accurate data by MALDI-TOF to represent the proportion or frequency of methylation at the CpG. In order to analyze the presence or absence of abnormal methylation of miR-34a promoter CpGs in osteosarcoma cells, we first detected and compared the methylation of miR-34a promoter CpGs in the osteoblast cell line (hFOB1.19) and osteosarcoma cell line (MG-63, SAOS-2), normal tissue specimens (n=12), and osteosarcoma tissue specimens (n=6) using MALDI-TOF (Figure 1A). Compared with hFOB1.19, the average methylation levels of the miR-34a promoter CpGs in MG-63 and SAOS-2 were higher (Figure 1B, P<0.05). Further analysis showed that the degrees of methylation of CpGs (CpG1, CpG2, CpG3, CpG4, CpG5, CpG8, CpG9, CpG10, CpG11, and CpG12.13) in the MG-63 and SAOS-2 were higher than those of hFOB1.19 (Figure 1C, P<0.05). The mean methylation level of the miR-34a promoter CpGs in the osteosarcoma tissue was also higher than that in the normal bone tissue, but the difference was not statistically significant (Figure 1D, P>0.05). Further analysis of the methylation of specific CpGs in the miR-34a promoter region showed that the degrees of methylation of miR-34a promoters, CpG1, CpG3, CpG5 and CpG7, in the osteosarcoma tissue were higher than those in the normal bone tissue (Figure 1E, P<0.05).
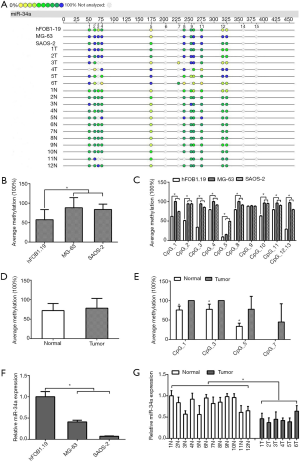
To determine whether the abnormal methylation of miR-34a promoters is associated with the expression of miR-34a in osteosarcoma cells or not, we detected and compared hFOB1.19 and MG-63, SAOS-2 using qPCR. Compared with hFOB1.19, the levels of miR-34aexpression were significantly lower in MG-63 and SAOS-2 (Figure 1F, P<0.05). We further detected and compared the normal bone tissue and the osteosarcoma tissue using qPCR and found that the mean miR-34a expression level was significantly lower in the osteosarcoma tissue than in the normal bone tissue (Figure 1G, P<0.05). This result was consistent with that in our experiment using osteosarcoma cells. All these results suggested that the abnormally low expression of miR-34a in osteosarcoma cells was related to the abnormal hypermethylation of the promoters, CpG1, CpG3, CpG5 and CpG7.
Low miR-34a expression was caused by the abnormal hypermethylation of miR-34a promoter CpGs in osteosarcoma cells
To investigate whether or not abnormal hypermethylation of the promoter induces decreased miR-34a expression and further promotes the proliferation of osteosarcoma cells, we used the MTT assay for MG-63 and SAOS-2 after 120 hours of culture in a medium added with DNA methyltransferase inhibitor 5-aza-dC, and drew the cell proliferation curve, the series of results are shown in Figure 2. The results showed that the proliferation rate of osteosarcoma cells was significantly inhibited in the 5-aza-dC culture group compared with the blank interference culture group (Figure 2A,B). We used the MALDI-TOF technique to detect MG-63 and SAOS-2 in the 5-aza-dC culture group and the blank interference culture group (Figure 2C). The methylation levels of miR-34a promoter CpGs in MG-63 and SAOS-2 were lower in the 5-aza-dC culture group, but the differences were not statistically significant (Figure 2D). We also analyzed DNMT1 using qPCR. The result showed that DNMT1 expression levels of MG-63 and SAOS-2 were increased in the 5-aza-dC culture group (Figure 2G, P>0.05), and this might be the reason that 5-aza-dC could not significantly reduce the methylation of the miR-34a promoter CpGs. We further analyzed the states of methylation in various miR-34a promoter CpGs, and found that the methylation levels of CpG1, CpG5 and CpG10 were decreased significantly in MG-63 (Figure 2E, P>0.05), while the methylation levels of CpG2, CpG5, and CpG12.13 were decreased significantly in SAOS-2 (Figure 2F, P>0.05).
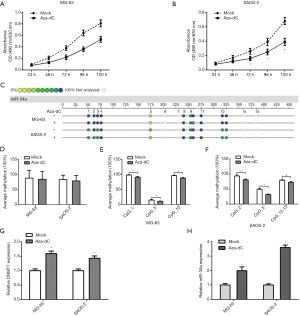
The expression levels of miR-34a in MG-63 and SAOS-2 were measured after MG-63 and SAOS-2 were cultured in a medium containing 5-aza-dC. The result showed that the miR-34a expression levels of two osteosarcoma cells in the 5-aza-dC culture group were increased significantly (Figure 2H, P>0.05). These results suggest that methylation of the miR-34a promoter CpGs may increase the expression of miR-34a and further inhibit the proliferation of osteosarcoma cells. Among them, CpG5 in the miR-34a promoter region was obviously representative. Also, 5-aza-dC increased the expression of DNMT1 in osteosarcoma cells and further positively regulated the methylation level of miR-34a promoter CpGs.
MiR-34a in osteosarcoma cells further negatively regulates the methylation of promoter CpGs by inhibiting DNMT1 expression
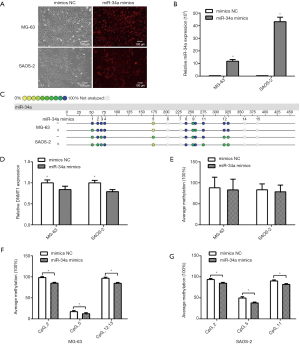
To study the regulatory effect of miR-34a on the methylation of promoter CpGs, we carried out transfection of miR-34a mimics for MG-63 and SAOS-2 to increase the miR-34a expression level as shown in Figure 3. After transfection with Cy3-labeled small RNAs, most osteosarcoma cells had fluorescent signals, and the signals were relatively strong, suggesting the efficiency of transfection was excellent (Figure 3A). After transfection, it was detected that the levels of miR-34a expression were increased significantly in two osteosarcoma cells (Figure 3B, P>0.05), and the mean methylation level of miR-34a promoter CpGs was decreased, but the difference was not statistically significant (Figure 3E, P<0.05). We further analyzed changed CpGs in the miR-34a promoter region of transfected miR-34a mimics in the osteosarcoma cell line (Figure 3C). The result showed that the methylation levels of CpG1, CpG5, and CpG12.13 were decreased significantly in MG-63 (Figure 3F, P>0.05), while the methylation levels of CpG2, CpG5 and CpG11 were significantly decreased in SAOS-2 (Figure 3G, P>0.05). The results suggested that miR-34a can reduce the methylation of its promoter CpGs. In order to investigate whether miR-34a underwent this process by regulating the methyltransferase gene DNMT1, we detected DNMT1 expression in the MG- 63 and SAOS-2 cell lines with transfected miR-34a mimics. The result showed that the expression of DNMT1 was decreased after the expression of miR-34a was increased (Figure 3D, P>0.05). It should be mentioned that the miR-34a promoter CpG5 is similarly representative.
Discussion
Currently, it is believed that osteosarcoma is caused by gene mutation during growth and development. There is much evidence that miRNA expression profiles in osteosarcoma are significantly specific, and some of these miRNAs are associated with the proliferation and metastasis of osteosarcoma (17,18). As one of these miRNAs, miR-34a has been studied extensively in osteosarcoma and other malignancies. Mutation of tumor suppressor gene p53 reduces the stimulation of miR-34a transcription, which affects the expression of miR-34a, and this type of regulation has been found in some osteosarcoma studies (19,20). In-vitro and in-vivo studies of osteosarcoma have shown that miR-34a can inhibit the growth and metastasis of osteosarcoma cells (10). Therefore, miR-34a can be considered as one of the tumor suppressor genes of osteosarcoma.
Abnormal hypermethylation of the promoter CpGs has been thought to be the leading cause of low miRNAs expression (21). Vogt et al. (12) found decreased miR-34a expression in colorectal cancer, kidney cancer, pancreatic cancer, breast cancer, uterine cancer, and soft tissue sarcoma, which affected the sensitivity of tumor to chemotherapy drugs. These authors also found that abnormal methylation of the promoter CpGs was an important reason for the decrease in the expression of miR-34a in these tumors. Moreover, miRNA participates in the transcription and regulation of genes by affecting the methylation of promoter CpGs (22). However, the promoter region of miR-34a contains multiple CpGs, while the osteosarcoma-specific abnormal CpG, the mutual regulation between it and miR-34a, and the related mechanism are not completely clear.
A number of studies have shown that the expression of miR-34a in the tumor tissue of patients with osteosarcoma is lower than that of normal adjacent tissues (23). It is well known that the mutation of genes in the normal human body is an important cause of oncogenesis. However, the difference in the expression of miR-34a between osteosarcoma and normal bone tissue is not clear. In the present study, we showed that the expression of miR-34a in the bone tissue of patients with osteosarcoma was lower than that in normal bone tissue. In order to further investigate the mechanism of low miR-34a expression in osteosarcoma cells, we used a more accurate MassARAAY technique, which can detect 500 base pair (bp) of nucleotides, including the methylation of 14 CpGs of the gene promoter (24). We found that compared with osteoblast cells and normal human bone tissue, miR-34a promoter CpG1, CpG3, CpG5, and CpG7 in the osteosarcoma cell lines and tissues were abnormally hypermethylated. It is generally believed that promoter hypermethylation can block the binding of transcription factors and thus prevent transcription and mRNA expression. Our results showed that there were only a few abnormally hypermethylated CpGs in the miR-34a promoter region, which were negatively correlated with the expression of the suppressor gene miR-34a, and these key CpGs could be important targets.
Recently, it has been reported that the methylation of miR-34b/c promoter CpGs in the miR-34s family is associated with the potential for tumor metastasis (25). In this study, proliferation of osteosarcoma cells was obviously inhibited after demethylation by 5-aza-dC, and in particular, methylation levels of the miR-34a promoter CpG1, CpG2, CpG5, CpG10, and CpG12.13 were decreased. This suggested that the hypermethylation environment might be one of the important reasons leading to highly invasive and metastatic properties of osteosarcoma.
We found that the expression of miR-34a was increased significantly in osteosarcoma cells treated by demethylation. It was confirmed that abnormal hypermethylation of some miR-34a promoter CpGs in the osteosarcoma cell could inhibit the expression of miR-34a. Additionally, we observed that 5-aza-dC could increase the expression of DNMT1 in osteosarcoma cells. Since DNMT1 was closely associated with the methylation and aggregation of tumor-associated gene CpGs, 5-aza-dC could competitively inhibit DNMT activity (26), which might lead to an increased expression of DNMT1. Though this process should be further verified, this phenomenon might explain the fact that 5-aza-dC could not reduce the methylation of all gene promoter CpGs and even increased the methylation of some CpGs in the present study and others (24).
The regulation of miR-34a on the methylation of promoter CpGs in osteosarcoma cells should be another aspect of this study. We increased miR-34a expression levels in the MG-63 and SAOS-2 cell lines and observed that the increase in miR-34a expression reduced the expression of DNMT1 and further negatively regulated the methylation of some CpGs, in which the state of methylation was significantly changed in CpG5. Previous studies suggested that DNMT1 in malignant tumors could inhibit tumor suppressor gene translation and promote tumorigenesis (16). Thus, we believe that miR-34a not only directly inhibits tumors, but also reduces the inactivation of other tumor suppressor genes by downregulating the expression of DNMT1.
The low expression of miR-34a has been reported as a predictor of postoperative recurrence of non-small cell carcinoma (27). During tumor development, DNA was released into the body fluids, and thus the methylation of miR-34a or miR-34b/c promoter CpGs in the bodily fluid could be used as a tumor diagnostic marker (28). In the present study, we found that some miR-34a promoter CpGs in the osteosarcoma tissue were abnormally hypermethylated compared with the normal human bone tissue, and the methylation of CpG5 was relatively active during the demethylation and increased expression of miR-34a. We also found that only a few CpGs played an important role in gene regulation. Later, experiments about the targeted intervention should be carried out for osteosarcoma based on the methylation characteristics of miR-34a promoter CpG5.The targeted diagnosis and treatment could be performed by utilizing the mutual regulation between miR-34a and promoter CpG5 methylation.
Conclusions
In this study, we confirmed that the hypermethylation of some specific miR-34a promoter CpGs in osteosarcoma cells decreased the expression of miR-34a. The increased expression of miR-34a could inhibit osteosarcoma DNMT1, and thus negatively regulated the methylation of miR-34a promoter CpGs. CpG5 was a representative promoter in the process of mutual regulation between miR-34a and promoter methylation. It is expected that CpG5 becomes the target of diagnosis and treatment of osteosarcoma.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.47). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the General Hospital of Ningxia Medical University. All issue specimens were collected after informed consent was obtained from each subject, and all specimens were treated according to legal standards for ethical handling and patient anonymity.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Berman SD, Calo E, Landman AS, et al. Metastatic osteosarcoma induced by inactivation of rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A 2008;105:11851-6. [Crossref] [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [Crossref] [PubMed]
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some micrornas downregulate large numbers of target mrnas. Nature 2005;433:769-73. [Crossref] [PubMed]
- Bommer GT, Gerin I, Feng Y, et al. P53-mediated activation of mirna34 candidate tumor-suppressor genes. Curr Biol 2007;17:1298-307. [Crossref] [PubMed]
- Chang TC, Wentzel EA, Kent OA, et al. Transactivation of mir-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007;26:745-52. [Crossref] [PubMed]
- Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: Mir-34a is a p53 target that induces apoptosis and G1-arrest. Cell cycle 2007;6:1586-93. [Crossref] [PubMed]
- Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 2008;582:1564-8. [Crossref] [PubMed]
- Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A 2007;104:15472-7. [Crossref] [PubMed]
- Wu X, Zhong D, Gao Q, et al. MicroRNA-34a inhibits human osteosarcoma proliferation by downregulating ether à go-go 1 expression. Int J Med Sci 2013;10:676-82. [Crossref] [PubMed]
- Yan K, Gao J, Yang T, et al. Microrna-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PloS one 2012;7:e33778. [Crossref] [PubMed]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597-610. [Crossref] [PubMed]
- Vogt M, Munding J, Grüner M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch 2011;458:313-22. [Crossref] [PubMed]
- Hatada I. The epigenomics of cancer. An omics perspective on cancer research. Springer, 2010:51-67.
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer-a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107. [Crossref] [PubMed]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253-66. [Crossref] [PubMed]
- Zhang Y, Chen FQ, Sun YH, et al. Effects of dnmt1 silencing on malignant phenotype and methylated gene expression in cervical cancer cells. J Exp Clin Cancer Res 2011;30:98. [Crossref] [PubMed]
- Sarver AL, Phalak R, Thayanithy V, et al. S-MED: sarcoma microRNA expression database. Lab Invest 2010;90:753-61. [Crossref] [PubMed]
- Miao J, Wu S, Peng Z, et al. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol 2013;34:2093-8. [Crossref] [PubMed]
- He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130-4. [Crossref] [PubMed]
- Wunder JS, Gokgoz N, Parkes R, et al. TP53 mutations and outcome in osteosarcoma: a prospective, multicenter study. J Clin Oncol 2005;23:1483-90. [Crossref] [PubMed]
- Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta 2010;1799:694-701. [Crossref] [PubMed]
- Huang J, Wang Y, Guo Y, et al. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus–related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010;52:60-70. [Crossref] [PubMed]
- He C, Xiong J, Xu X, et al. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun 2009;388:35-40. [Crossref] [PubMed]
- Li H, Yu G, Shi R, et al. Cisplatin-induced epigenetic activation of miR-34a sensitizes bladder cancer cells to chemotherapy. Mol Cancer 2014;13:8. [Crossref] [PubMed]
- Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008;105:13556-61. [Crossref] [PubMed]
- Li Q, Bartlett DL, Gorry MC, et al. Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol Pharmacol 2009;76:1072-81. [Crossref] [PubMed]
- Gallardo E, Navarro A, Viñolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903-9. [Crossref] [PubMed]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. New Engl J Med 2008;358:1118-28. [Crossref] [PubMed]


