
Helicase lymphoid-specifics and selenoprotein P1 are potential candidate genes in the progression and prognosis of lung adenocarcinoma
IntroductionOther Section
At present, lung cancer remains to be one of the most common malignant tumors and leads to increasing mortality around the world (1,2). During the past decades, the occurrence and mortality of lung cancer are rising rapidly worldwide, especially in those countries with advanced industry (3). Lung adenocarcinoma (AD) is the main histologic subtype of lung carcinoma (4). Although numerous studies verify multiple oncogenes participating in the pathogenesis of lung cancer, the overall 5-year survival rate of lung patients is still very lower (4,5). Thus, identification of molecular mechanisms and pathways for the development of lung tumors will benefit to the therapeutic effect and improve the outcomes of lung AD patients.
High throughput technology provides promising methods for cancer research, such as the molecular diagnosis and prognosis prediction of cancer (6,7). Previous studies have reported numerous potential biomarkers and therapeutic targets of lung cancer through microarrays or sequencing data (8). Girard et al. developed an mRNA expression signature including 62 genes to discriminate non-small cell lung cancer (NSCLC) patients. The researchers demonstrated high predictive accuracies (93–95%) had been obtained in the TCGA and other public database using their signature (9). Meanwhile, Chen et al. developed a malignancy-risk gene signature that is significant related to overall survival (OS) of NSCLC patients. This signature could be used for the early identification of NSCLC patients (10). Therefore, microarray data provides us new opportunities to identify novel target genes and construct prediction models for lung cancer, which may promote the treatments of lung cancer patients.
In our current study, we downloaded original microarray data from Gene Expression Omnibus (GEO) to screen differentially expressed genes (DEGs) between AD and normal controls. Functional levels of DEGs were subsequently performed through gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Moreover, protein-protein interaction (PPI) network was performed to pick out hub genes in the developments of AD. In addition, prognostic effects of hub genes were evaluated in the Kaplan Meier plotter and GEPIA databases. We hoped to have an in-deep understanding of the pathogenesis of AD, then to identify potential candidate targets or biomarkers for the early prediction and diagnosis of AD patients.
MethodsOther Section
Data processing of DEGs
In our study, GSE43458 gene expression profiles were extracted from the GEO dataset. GSE43458, submitted by Kabbout et al. (11), was performed by GPL6244 platform (Affymetrix Human Gene 1.0 ST Array). Only never smoking (n=40) AD samples and 30 normal tissues were included in our current study. Limma package was adopted to screen the DEGs between normal tissues and AD samples. |log2foldchange (FC)| >1 was considered as the threshold to determine the significant difference of gene expression. GO and KEGG pathway analysis were carried out to investigate the DEGs at the functional level using the Database for Annotation, Visualization and Integrated Discovery (DAVID). False discovery rate (FDR) <0.05 was set as significantly difference.
PPI network analysis
The Search Tool for the Retrieval of Interacting Gene (STRING) database was performed to construct interaction network among DEGs (12). Combined score more than 0.4 was defined as the threshold. What’s more, the network was visualized by Cytoscape software. The MCODE plug of Cytoscape was applied to display the major modules of PPI network. The criteria were considered as following: MCODE score ≥10 and number of nodes ≥4. MCODE was based on vertex weighting by local neighborhood density and outward traversal from a locally dense seed protein to isolate the dense regions according to given parameters (13). The algorithm identified seed nodes for expansion by computing a score of local density for each node in the graph. The algorithm expands highly scoring seed nodes in a local search procedure by adding highly scoring nodes connected to the module (14).
Validation of the hub genes
The Oncomine database (www.oncomine.org) includes 500 cancer types with gene expression and sample data. Currently, more than 490 datasets and nearly 40,000 measured samples were contained in the Oncomine database. According to a large amount of data, the Oncomine provides several analytical tools, including differential expression analysis, co-expression analysis, and comparing analysis. To validate the dysregulation of the hub genes, we applied the Oncomine database to confirm the differentially expression levels of hub genes. In our current study, the expression levels of cancer and normal control were compared with the Students’ t-test. The P value was established at 1×10−4 and FC was defined as 2. At the same time, the data type was limited to mRNA. Moreover, GEPIA dataset (http://gepia.cancer-pku.cn/) (15), another public database, was also used to explore the differential expression of hub genes.
Survival analysis of hub genes
The Kaplan Meier plotter database (www.kmplot.com), was applied to evaluate the prognostic values of hub genes (16). Up to now, 2,437 lung cancer patients were included in this database. Depending on the expression levels (high vs. low) of hub genes, patients were divided into two groups. We analyzed the survival time of AD patients using a Kaplan-Meier survival plot. The hazard ratio (HR) with 95% confidence intervals and log rank P value were calculated and displayed on the plot. Meanwhile, the effect of hub genes on the prognosis of lung AD patients were validated in the GEPIA database.
Statement of ethics approval
Our study was investigated using public database. Participants had been giving informed consent before taking part in the Oncomine, GEPIA and Kaplan Meier plotter databases. Moreover, this study had got approval from the Institutional Ethics Review Board of Affiliated Hospital of Jining Medical University (ID: 20181026B).
ResultsOther Section
Gene expression profiling in AD
In our current study, a comparative analysis between AD samples and normal lung tissues identified 589 DEGs (278 up-regulated genes and 581 down-regulated genes) according to the cut-off criteria (FDR <0.05 and log |FC|>1), as shown in Figure 1A. The heat map of top 100 genes was displayed in Figure 1B.
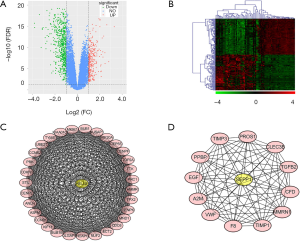
GO and KEGG pathway enrichment analysis
All DEGs were uploaded to the online DAVID database to identify the most significantly GO terms and KEGG pathways. GO results suggested DEGs were markedly related to biological processes, including cell adhesion, regulation of cell migration and regulation of cell motility (Table 1). For molecular function, the DEGs were involved in calcium ion binding, glycosaminoglycan binding and carbohydrate binding (Table 1). For cellular component, the DEGs were belonged to extracellular space, extracellular region, and extracellular region part (Table 1). KEGG analysis suggested the most significant pathways of the DEGs were extracellular matrix (ECM)-receptor interaction, complement and coagulation cascades, and vascular smooth muscle contraction (Table 2).
Table 1
| GO | Pathway description | Gene count | FDR |
|---|---|---|---|
| Process | |||
| GO.0007155 | Cell adhesion | 94 | 5.23E-17 |
| GO.0030334 | Regulation of cell migration | 67 | 4.03E-14 |
| GO.2000145 | Regulation of cell motility | 68 | 1.83E-13 |
| GO.0051270 | Regulation of cellular component movement | 72 | 3.00E-13 |
| GO.0051239 | Regulation of multicellular organismal process | 151 | 5.61E-13 |
| Function | |||
| GO.0005509 | Calcium ion binding | 55 | 4.13E-05 |
| GO.0005539 | Glycosaminoglycan binding | 25 | 5.29E-05 |
| GO.0030246 | Carbohydrate binding | 30 | 6.90E-05 |
| GO.0005515 | Protein binding | 204 | 0.00765 |
| GO.0005102 | Receptor binding | 65 | 0.00986 |
| Component | |||
| GO.0005615 | Extracellular space | 133 | 5.80E-29 |
| GO.0005576 | Extracellular region | 271 | 1.68E-23 |
| GO.0044421 | Extracellular region part | 236 | 1.50E-21 |
| GO.0005886 | Plasma membrane | 262 | 3.63E-19 |
| GO.0071944 | Cell periphery | 265 | 3.63E-19 |
GO, gene ontology; DEG, differentially expressed gene; FDR, False discovery rate; AD, lung adenocarcinoma.
Table 2
| Pathway ID | Pathway description | Gene count | FDR |
|---|---|---|---|
| 4512 | ECM-receptor interaction | 14 | 0.000596 |
| 4610 | Complement and coagulation cascades | 12 | 0.000757 |
| 4270 | Vascular smooth muscle contraction | 15 | 0.00167 |
| 4514 | Cell adhesion molecules (CAMs) | 16 | 0.00196 |
| 5144 | Malaria | 9 | 0.00196 |
KEGG, Kyoto Encyclopedia of Gene and Genome; DEG, differentially expressed gene; FDR, False discovery rate; ECM, extracellular matrix; AD, lung adenocarcinoma.
PPI network analysis of DEGs
Based on the information in the STRING database, PPI network was presented by Cytoscape software. According to the MCODE score, only two modules were with more than 10 scores. Especially, helicase lymphoid-specific (HELLs) and selenoprotein P1 (SEPP1) were considered as the seed genes of two modules, which might function as key genes for lung AD occurrence and development (Figure 1C,D). In the GSE43458 dataset, HELLs upregulated 2.05 folds and SEPP1 downloaded 2.01 folds in the AD patients.
Different transcription levels of HELLs and SEPP1
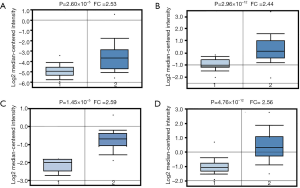
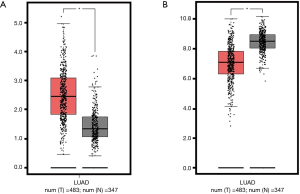
Up to now, four studies showed AD patients had higher HELLs expression in Oncomine database, compared with normal controls. In Su’s dataset, HELLs mRNA level increased 2.53 folds (P=2.60×10−5) in the AD samples (Figure 2A). Okayama’s dataset revealed HELLs mRNA level increased 2.44 folds (P=2.96×10−12) in the AD samples (Figure 2B). In Garber’s dataset, the transcription levels of HELLs up-regulated 2.59 folds in the lung AD (P=1.45×10−5) (Figure 2C). The HELLs mRNA level also increased 2.56 folds in the Hou’s AD group (P=4.76×10−12), compared with normal samples (Figure 2D). In addition, 483 AD patients and 347 normal controls were included to compare the expression levels of HELLs mRNA in the GEPIA database. The results also revealed HELLs mRNA level increased in the AD patients (P<0.01) (Figure S1A).
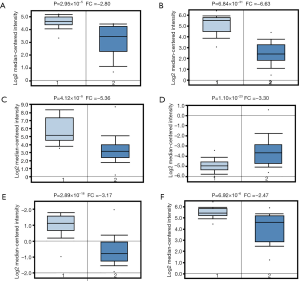
There were six studies revealing the downregulated SEPP1 in the Oncomine database. In Stearman’s study, SEPP1 mRNA level decreased 2.80 folds in AD patients, compared with normal tissues (P=2.95×10−5) (Figure 3A). In Selamat’s study, SEPP1 mRNA level decreased 6.63 folds in AD patients (P=6.84×10−31) (Figure 3B). Compared with normal lung tissues, Bhattacharjee’s study indicated SEPP1 mRNA level decreased 5.36 folds (P=4.12×10−6) (Figure 3C). Beer’s study suggested SEPP1 mRNA level decreased 3.30 folds (P=1.10×10−23) in the AD patients (Figure 3D). In Hou’s study, SEPP1 mRNA level decreased 3.17 folds (P=2.89×10−18), compared with normal lung samples (Figure 3E). Moreover, Su’s study suggested SEPP1 mRNA level decreased 2.47 folds (P=6.92×10−6) in AD patients (Figure 3F), when compared with normal lung tissues. Meanwhile, our results also suggested SEPP1 mRNA level decreased in the 483 AD patients, when compared with 347 normal controls in the GEPIA database (P<0.01) (Figure S1B).
The prognostic effect of hub genes for the lung AD patients
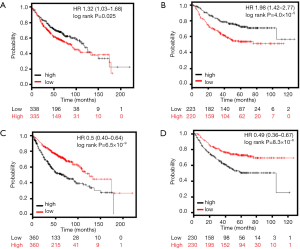
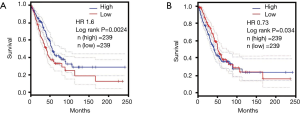
We explored the prognostic value of HELLs in the Kaplan Meier plotter and GEPIA databases. In lung AD patients, Kaplan-Meier analysis results indicated high-HELLs patients exhibited shorter OS periods than low-HELLs patients [HR 1.32 (1.03–1.68), P=0.025] (Figure 4A). Kaplan-Meier analysis also indicated that high-HELLs exhibited shorter progression-free survival (PFS) periods than low-HELLs patients [HR 1.98 (1.42–2.77), P=4.0×10−5] (Figure 4B). GEPIA database results also showed AD patients with higher HELLs exhibited shorter OS periods than those with lower HELLs (HR 1.6, P=0.0024) (Figure S2A). Meanwhile, prognostic value of the SEPP1 was also evaluated by Kaplan-Meier analysis. Kaplan-Meier analysis suggested low-SEPP1 AD patients had shorter OS periods than high-SEPP1 patients [HR 0.5 (0.40–0.64), P=6.5×10−9] (Figure 4C). Results also demonstrated patients with low-SEPP1 exhibited shorter PFS periods than patients with high-SEPP1 [HR 0.49 (0.36–0.67), P=8.3×10−6] in lung AD patients (Figure 4D). In addition, GEPIA database results also showed low-SEPP1 patients exhibited shorter OS periods than high-SEPP1 patients (HR 0.73, P=0.034) (Figure S2B)
DiscussionOther Section
Although the occurrence of lung cancer has declined, it still results in the majority of cancer deaths all over the world, which is mainly caused by the failure of early prediction and diagnosis of lung cancer patients. Therefore, it was urgently important to explore the deep pathogenesis of lung cancer. In this study, we compared the gene expression between 40 AD samples and 30 normal tissues in GSE43458. Totally, 859 DEGs were identified, including 278 up-regulated genes and 581 down-regulated genes. To deepen our understanding of those DEGs, GO function and KEGG pathway analysis were carried out. Our results demonstrated those DEGs were involved in cell adhesion, migration and motility. Moreover, upregulated HELLs and downregulated SEPP1 were considered as hub genes in the progress of AD. Furthermore, prognostic analysis results also indicated upregulated HELLs and downregulated SEPP1 were associated with the survival time of AD patients.
HELLs is one of members of the SNF2 (Sucrose Non-Fermenter) family of helicase proteins, which contribute to chromatin recombination, remodeling, transcription and methylation (17). High levels of HELLs have been identified in several human cancers, including leukemia, NSCLC, breast cancer, and melanoma (18). Waseem et al. demonstrated HELLs mRNA and protein expression were significantly correlated with the progression of head and neck squamous cell carcinoma. This study provided evidence that HELLs might be hub gene for early cancer discrimination and indicator of malignant conversion and progression. Moreover, the high expression of HELLs was validated in renal cell carcinoma. High mRNA level of HELLs was an independent predictor of poor outcome in renal cell carcinoma patients (19). Yano et al. suggested downregulation of HELLs by allelic loss and abnormal proteins produced by tumors specific exon might result in malignancy or progression of lung cells (20). Furthermore, HELLs activity was considered as a critical component of the malignant progression of cancer, and HELLs could be a promising therapeutic target for cancer patients (21).
SEPP1 gene, with 10 selenocysteine residues, contributes to the selenium transport and production of other selenoproteins (22). Previous studies demonstrated SEPP1 played an important role in cancer prevention through its function in mediating oxidative damage (23,24). When the SEPP1 gene reduced, the oxidative stress increased and resulted in carcinogenesis (25). At present, the downregulation of plasma SEPP1 in various cancers has been established, including prostate and colorectal cancer (26,27). Gresner’s study (28) suggested SEPP1 gene expression downregulated in malignant NSCLC lung tissue, compared with paired non-malignant control tissue. In our study, we found SEPP1 was downregulated in the AD patients, which was consistent with previous reports (26,27). However, those studies about the deeply mechanism of SEPP1 to affect the development of lung cancer were limited.
Our study had several attributes that strengthen its validity. Using bioinformatics methods, we analyzed the DEGs from the global gene levels, and identified the important roles of HELLs and SEPP1 in the development of lung AD. Dysregulation of HELLs and SEPP1 and their clinical effects were validated in large online databases. However, some limitations also were obvious. First, AD just represents one subgroup of lung cancer, so we do not analyze the DEGs in patients with different subtype, such as lung squamous cell carcinoma and small cell lung cancer. Therefore, the HELLs and SEPP1 might have a variety of functions on the progression of different lung cancer subgroups. Second, although we validated the dysregulation of HELLs and SEPP1 mRNA in the Oncomine and GEPIA database, the protein levels of HELLs and SEPP1 were not studied in our study. Moreover, further studies were needed to deeply investigate the mechanisms of HELLs and SEPP1 affecting the development of lung cancer.
ConclusionsOther Section
Our current study performed a comprehensive analysis of DEGs contributing to the progress of AD. HELLs and SEPP1 were identified as core genes of AD. However, further experiments were needed to investigate the molecular biological function.
AcknowledgmentsOther Section
We acknowledge Dr. Don Ma and Yan Zhang for data analysis and manuscript writing.
Funding: None.
FootnoteOther Section
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.45). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study had got approval from the Institutional Ethics Review Board of Affiliated Hospital of Jining Medical University (ID: 20181026B). Participants had been giving informed consent before taking part in the Oncomine, GEPIA and Kaplan Meier plotter databases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Yuan S, Liu Q, Hu Z, et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis 2018;9:450. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Zhou J, Xiao H, Yang X, et al. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep 2018;8:37. [Crossref] [PubMed]
- Azzawi H, Hou J, Xiang Y, et al. Lung cancer prediction from microarray data by gene expression programming. IET Syst Biol 2016;10:168-78. [Crossref] [PubMed]
- Shukla S, Evans JR, Malik R, et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J Natl Cancer Inst 2016; [Crossref] [PubMed]
- Choi H, Na KJ. A Risk Stratification Model for Lung Cancer Based on Gene Coexpression Network and Deep Learning. Biomed Res Int 2018;2018:2914280. [Crossref] [PubMed]
- Girard L, Rodriguez-Canales J, Behrens C, et al. An Expression Signature as an Aid to the Histologic Classification of Non-Small Cell Lung Cancer. Clin Cancer Res 2016;22:4880-9. [Crossref] [PubMed]
- Chen DT, Hsu YL, Fulp WJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011;103:1859-70. [Crossref] [PubMed]
- Kabbout M, Garcia MM, Fujimoto J, et al. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res 2013;19:3383-95. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Rivera CG, Vakil R, Bader JS. NeMo: Network Module identification in Cytoscape. BMC Bioinformatics 2010;11:S61. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241. [Crossref] [PubMed]
- Raabe EH, Abdurrahman L, Behbehani G, et al. An SNF2 factor involved in mammalian development and cellular proliferation. Dev Dyn 2001;221:92-105. [Crossref] [PubMed]
- Waseem A, Ali M, Odell EW, et al. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol 2010;46:536-42. [Crossref] [PubMed]
- Chen D, Maruschke M, Hakenberg O, et al. TOP2A, HELLS, ATAD2, and TET3 Are Novel Prognostic Markers in Renal Cell Carcinoma. Urology 2017;102:265.e1-e7. [Crossref] [PubMed]
- Yano M, Ouchida M, Shigematsu H, et al. Tumor-specific exon creation of the HELLS/SMARCA6 gene in non-small cell lung cancer. Int J Cancer 2004;112:8-13. [Crossref] [PubMed]
- von Eyss B, Maaskola J, Memczak S, et al. The SNF2-like helicase HELLS mediates E2F3-dependent transcription and cellular transformation. EMBO J 2012;31:972-85. [Crossref] [PubMed]
- Traulsen H, Steinbrenner H, Buchczyk DP, et al. Selenoprotein P protects low-density lipoprotein against oxidation. Free Radic Res 2004;38:123-8. [Crossref] [PubMed]
- Burk RF, Hill KE, Motley AK. Selenoprotein metabolism and function: evidence for more than one function for selenoprotein P. J Nutr 2003;133:1517S-20S. [Crossref] [PubMed]
- Sasakura C, Suzuki KT. Biological interaction between transition metals (Ag, Cd and Hg), selenide/sulfide and selenoprotein P. J Inorg Biochem 1998;71:159-62. [Crossref] [PubMed]
- Short SP, Whitten-Barrett C, Williams CS. Selenoprotein P in colitis-associated carcinoma. Mol Cell Oncol 2015;3:e1075094. [Crossref] [PubMed]
- Al-Taie OH, Uceyler N, Eubner U, et al. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer 2004;48:6-14. [Crossref] [PubMed]
- Calvo A, Xiao N, Kang J, et al. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res 2002;62:5325-35. [PubMed]
- Gresner P, Gromadzinska J, Jablonska E, et al. Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer 2009;65:34-40. [Crossref] [PubMed]


