
Noninvasive circulating tumor cell and urine cellular XPC (rs2228001, A2815C) and XRCC1 (rs25487, G1196A) polymorphism detection as an effective screening panel for genitourinary system cancers
Introduction
Genitourinary system cancers, such as bladder cancer, renal carcinoma, and prostate cancer, frequently occur in males. In 2019, there were 61,700, 44,120, and 174,650 new cases of bladder cancer, renal carcinoma, and prostate cancer, respectively, in American males and 18,770, 29,700, and 0 female patients with bladder cancer, renal carcinoma, and prostate cancer, respectively (1). This incidence trend was consistent with that in China (2). Although these patients undergo timely surgical resection, the cancer is likely to recur, and thus long-term monitoring is needed. Therefore, it is crucial to choose highly sensitive and noninvasive indicators to screen and monitor the progression of genitourinary system cancers.
Circulating tumor cells (CTCs), which originate from carcinoma tissue, were first discovered in peripheral blood by the Australian physician Thomas Ashworth in 1869 (3). CTCs have been widely used in cancer screening and monitoring studies, including for breast cancer, small cell lung cancer, bladder cancer, prostate cancer, and other solid tumors (4-7). CTCs from 188 urothelial carcinomas of bladder (UCB) patients were analyzed by the CellSearch system, revealing that patients without CTCs had better relapse-free survival (RFS) than did patients with CTCs detected by histology (P<0.05) (8). In patients with advanced urothelial carcinoma of the bladder, CTCs have been associated with tumor stage, lymph node metastases, and fluorodeoxyglucose positron emission tomography (FDG-PET) findings (9). CTCs have also been used for sunitinib monitoring in metastatic renal cell carcinoma (mRCC), with very good results (10). When discriminating prostate cancer metastasis from nonmetastasis, glucose metabolic (GM) + CTCs presented a higher area under the receiver operating characteristic (ROC) curve (AUC), of 0.780, than did EMT CTC subtypes (E-CTCs 0.729, H-CTCs 0.741, and M-CTCs 0.648) (11). In addition, based on monitoring 6,081 patients, the efficacy of CTC was better than that of SPA in metastatic castration-resistant prostate cancer (12).
Many studies have verified tumor susceptibility genes related to the occurrence and development of cancers. In general, genitourinary system cancers have similar familial syndromes; for instance, hereditary renal cell carcinoma (RCC) exhibits germline mutations in VHL, FLCN, MET, FH, SDHB/C/D, and TSC1/2 (13). Geary et al. found a 3.6 relative risk for bladder cancer (P=0.001) in MutS homolog 2 (MSH2)-positive families compared with MSH2-negative families (14). Furthermore, mutations in EGFR, ERBB2, PTEN, APC, NF1, and BRAF have been identified in mRCC patients based on ctDNA detection, with a correlation with tumor burden (15). However, ctDNA is very easily degraded because of its short half-life of 16 min to 2.5 hours and is hence not an ideal biomarker (16,17). In a genome-wide association study (GWAS), 76 prostate cancer risk SNPs were found; the MYC, MSMB, KLK2 and KLK3 genes, which are clinically relevant, were analyzed with regard to prostate cancer risk (18). The XRCC1 polymorphism rs1799782 (C194T) was reported to correlate with tumor metastasis and molecular subtypes in breast cancer and XRCC1 Arg399Gln with bladder cancer (19,20). In addition, the XPC intron11 C/A polymorphism and Lys939Gln are risk factors for prostate cancer (21,22).
In the process of biomarker detection, different methods may have a considerable impact on detection results. The CellSearch system for CTCs, which is based on positive antibody capture, showed a low detection rate (23), whereas subtraction enrichment-immunofluorescence in situ hybridization (SE-iFISH) displayed better performance (SE-iFISH vs. CellSearch, 91% vs. 38%) (24). SE-iFISH involves subtraction enrichment of hematogenous cells and subsequent removal of red and white blood cells, leaving only CTCs (25). In addition to peripheral blood samples, noninvasive urine cells have been used in recurrence monitoring for bladder cancer, an approach that was completely harmless to patients (26).
To increase the sensitivity and specificity of genitourinary system cancer screening and to avoid causing more trauma to patients, we combined multiple detection methods in this study. SE-iFISH was employed to detect CTCs from peripheral blood samples, and Agena Bioscience MassARRAY was used to reveal mutations or SNPs in tumor susceptibility genes in urine cells. Agena Bioscience MassARRAY is an advanced detection system based on MALDI-TOF MS technology and has a high flux function for detecting dozens of genes in one sample (27). By using multiple detection methods, we hope to improve sensitivity and specificity for genitourinary system cancer screening.
Methods
Patients
Patients were enrolled from two hospitals, Quanzhou First Hospital Affiliated to Fujian Medical University (Fujian, China) and Xiangyang No. 1 People’s Hospital (Hubei, China), from May 2016 to Jan 2019. Permission to use peripheral blood and urine samples was obtained from the Ethics Committees of these two hospitals. The study enrolled 87 subjects, including 67 patients and 20 healthy individuals; 77 were male and 10 female, with average age of 61.15 years (Table 1). All patients and healthy individuals signed informed consent before the experiment. Peripheral blood specimens (7.5 mL each) were extracted from 36 patients for SE-iFISH CTCs detection, including 16 bladder cancer patients, 12 renal carcinoma patients, and 8 prostate cancer patients. Urine specimens (50 mL each) were collected from 65 subjects for MassARRAY detection, including 26 bladder cancer patients, 5 renal carcinoma patients, 4 prostate cancer patients, 4 benign bladder tumor patients, 6 prostatitis patients, and 20 healthy individuals.
Table 1
| Variable | No. of participants | % of participants | Average age | ||
|---|---|---|---|---|---|
| Men | Women | All | |||
| Bladder cancer patients | 28 | 6 | 34 | 39.08% | 67.20 |
| Renal carcinoma patients | 12 | 1 | 13 | 14.94% | 54.00 |
| Prostate cancer patients | 10 | 0 | 10 | 11.49% | 64.20 |
| Benign bladder tumor patients | 4 | 0 | 4 | 4.60% | 55.75 |
| Prostatitis patients | 6 | 0 | 6 | 6.60% | 55.50 |
| Healthy individuals | 17 | 3 | 20 | 22.99% | 56.75 |
SE-iFISH CTC enrichment and identification
Subtraction enrichment and identification reagents for analysis of CTCs were provided by the Cytelligen CTC enrichment kit (Cytelligen, San Diego, CA, USA). For CTC subtraction enrichment, briefly, peripheral blood (7.5 mL) was collected into acid citrate dextrose (ACD) anticoagulant tubes and centrifuged for 8 min at 800 ×g for plasma removal. The blood cells were transferred to a centrifuge tube containing 3 mL of the hCTC separation matrix and centrifuged for 8 min at 450 ×g to pellet the red blood cells. The buffy-coat cells were collected in tubes and incubated on horizontal rotators with an immunomagnetic particle-conjugated anti-CD45 antibody. The samples were shaken for 20 min at 200 ×g, and the tubes were placed on a magnetic stand (Axygen, IMAG-150-I-G, USA) to remove leukocytes and obtain CTCs. After cleaning, the CTCs were fixed on a glass slide and dried overnight.
For CTC identification, the dried cells were treated with 20 µL FR1 and 180 µL FR2 mixture for 10 min and washed twice with FR3. The cells were dehydrated with ethanol. After air drying, 10 µL of probe solution (fluorescence-labeled CEP8 probes) was added to the glass slide, and fluorescence in situ hybridization (FISH) with denaturation at 76 °C for 10 min and hybridization at 37 °C for 4 hours was performed. The cells were then immunostained with anti-CD45 and anti-CK18 antibodies for 2 hours at room temperature. After washing with PBS, the slide was treated with DAPI reagent and observed under a fluorescence microscope (Nikon, Japan). CTCs were confirmed when CK18+CD45-DAPI+CEP8 =2, CK18+CD45-DAPI+CEP8 >2, and CK18-CD45-DAPI+CEP8 >2.
MassARRAY SNP detection
MassARRAY iPLEX Gold multiple genotyping analysis (Agena Bioscience, Inc.) reagents (Agena PCR reagent, Agena SAP reagent, and Agena iPLEX reagent) were provided by Agena Bioscience. Briefly, 50 mL of urine was centrifuged 800 ×g for 10 min to obtain cells. DNA extraction was performed following the manufacturer’s protocol (TIANGEN, DP348). PCR mixtures were prepared following the Agena PCR reagent set and start PCR procedures, with shrimp alkaline phosphatase (SAP) treatment. After extending the reaction, the samples underwent desalination processing and were added to the chip and analyzed by MALDI-TOF MS.
MassARRAY primers:
- XPC (rs2228001, A2815C) F: ACGTTGGATGAGCAGCTTCCCACCTGTTC.
- R: ACGTTGGATGAACTGGTGGGTGCCCCTCTA.
- UEP: ACCTGTTCCCATTTGAG.
- XRCC1 (rs25487, G1196A) F: ACGTTGGATGCATCGTGCGTAAGGAGTGG.
- R: ACGTTGGATGCAGGATAAGGAGCAGGGTTG.
- UEP: CATGCGTCGGCGGCTGCCCTCCC.
Results
SE-iFISH CTCs had excellent sensitive and specific screening effects for genitourinary system cancers
To evaluate the sensitivity and specificity of SE-iFISH CTC detection for cancer screening, 10 healthy individuals and 10 treated patients with genitourinary system cancers (4 BC patients, 4 renal carcinoma patients, and 2 prostate cancer patients) were enrolled for SE-iFISH CTC analysis. Peripheral blood specimens (7.5 mL) were extracted from subjects, and serum, red blood cells, and white blood cells were removed; CTCs were retained for iFISH CTC detection. According to the results, 0 CTCs were detected in the 10 healthy individuals. In contrast, CTCs were found in 70% (7/10) of the patients, with an average number of 4.3/7.5 mL peripheral blood (Figure 1A,B). SE-iFISH CTC detection had very high specificity in distinguishing genitourinary system cancer patients from healthy individuals (Figure 1B).
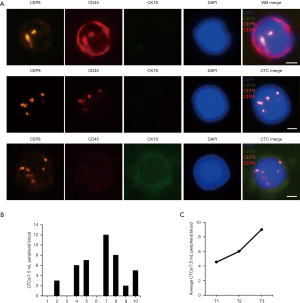
Next, SE-iFISH CTC detection was performed for 26 preoperative patients, including 12 bladder cancer, 8 renal carcinoma, and 6 prostate cancer patients. We found a CTC positivity rate of 76.92% (20/26), and the average number of CTCs was 5.5/7.5 mL peripheral blood per patient (Table 2). Further analysis revealed that the number of CTCs was consistent with progression of the cancer, with average CTC numbers in tumor stages T1, T2, and T3 of 4.53, 6.00, and 9.00, respectively (Figure 1C). Thus, CTC has high application value in genitourinary system cancer screening and may effectively reflect cancer progression.
Table 2
| No. | Sex | Disease | CTCs | Urine cytology | Tumor stage |
|---|---|---|---|---|---|
| 1 | Male | Bladder cancer | 6 | IV | T1N0M0 |
| 2 | Male | Bladder cancer | 4 | V | T1N0M0 |
| 3 | Male | Bladder cancer | 7 | V | T3bN1M0 |
| 4 | Male | Bladder cancer | 0 | IV | T1N0N0 |
| 5 | Female | Bladder cancer | 3 | IV | T1N0M0 |
| 6 | Male | Bladder cancer | 4 | T1N0M0 | |
| 7 | Female | Bladder cancer | 9 | T2bN0M0 | |
| 8 | Female | Bladder cancer | 7 | T2aN0M0 | |
| 9 | Male | Bladder cancer | 9 | T1N0M0 | |
| 10 | Male | Bladder cancer | 6 | T2bN0M0 | |
| 11 | Male | Bladder cancer | 0 | T2N1M0 | |
| 12 | Male | Bladder cancer | 0 | T1bN0M0 | |
| 13 | Male | Renal carcinoma | 9 | T1N0M0 | |
| 14 | Female | Renal carcinoma | 12 | T1N0M0 | |
| 15 | Male | Renal carcinoma | 3 | T1N0M0 | |
| 16 | Male | Renal carcinoma | 5 | T1bN0M0 | |
| 17 | Male | Renal carcinoma | 6 | T1N0M0 | |
| 18 | Male | Renal carcinoma | 0 | T1N0M0 | |
| 19 | Male | Renal carcinoma | 0 | T2aN0M0 | |
| 20 | Male | Renal carcinoma | 6 | T2aN0M0 | |
| 21 | Male | Prostate cancer | 12 | T2cN0M0 | |
| 22 | Male | Prostate cancer | 8 | T3bN1M0 | |
| 23 | Male | Prostate cancer | 12 | T3bN0M0 | |
| 24 | Male | Prostate cancer | 7 | T1N0M0 | |
| 25 | Male | Prostate cancer | 0 | T1N0M0 | |
| 26 | Male | Prostate cancer | 8 | T2bN0M0 |
CTC, circulating tumor cell.
XPC (rs2228001, A2815C) and XRCC1 (rs25487, G1196A) polymorphisms as an effective complement for CTC screening of genitourinary system cancers
Because urine samples are completely noninvasive and there is ample cancer information in the urine cells of patients (26), we sought to utilize a new method to study genitourinary system cancer screening by using these noninvasive samples. Many genes involved in genitourinary system cancers, such as XRCC1, XPC, EGFR, ERBB2, APC, TERT, and BRAF, have been studied by the advanced and high-flux MassARRAY iPLEX Gold multiple genotyping analysis system (15,19,21). After preliminary screening, we found that the XPC (rs2228001, A2815C) and XRCC1 (rs25487, G1196A) polymorphisms were effectively detected by this system, and these two gene polymorphisms had good specificity for distinguishing cancer patients from noncancer patients and healthy individuals.
A total of 65 subjects, including 26 bladder cancer patients, 5 renal carcinoma patients, 4 prostate cancer patients, 4 benign bladder tumor patients, 6 prostatitis patients, and 20 healthy individuals (Table 3), were evaluated, and we found that positivity rates for XPC A2815C (A>C) of 30.77%, 40%, and 50% among bladder cancer patients, renal carcinoma patients, and prostate cancer patients, respectively, with rates of 3.85%, 20%, 25% for XRCC1 G1196A (G>A) (Figure 2A). To our surprise, no samples from benign bladder tumor patients, prostatitis patients, and healthy individuals were positive for XPC A2815C and XRCC1 G1196A. Thus, XPC A2815C and XRCC1 G1196A have high specificity in differentiating genitourinary cancer patients from those without cancer.
Table 3
| Variable | No. of participants | % of XPC A2815C | % of XRCC1 G1196A | ||
|---|---|---|---|---|---|
| Men | Women | All | |||
| Bladder cancer patients | 21 | 5 | 26 | 30.77% | 3.85% |
| Renal carcinoma patients | 4 | 1 | 5 | 40% | 20% |
| Prostate cancer patients | 4 | 0 | 4 | 50% | 25% |
| Benign bladder tumor patients | 4 | 0 | 4 | 0% | 0% |
| Prostatitis patients | 6 | 0 | 6 | 0% | 0% |
| Healthy individuals | 17 | 3 | 20 | 0% | 0% |
SNP, single-nucleotide polymorphism.
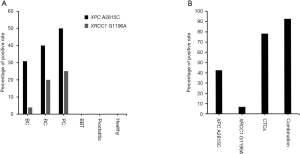
Despite their high specificity, we found a lower positive rate (50% and 25%) for XPC A2815C and XRCC1 G1196A (Figure 2A) when compared to that of CTCs (76.92%) (Table 2). Therefore, we combined these two detection methods to explore new detection modes. For 14 patients (8 bladder cancer, 4 renal carcinoma, and 2 prostate cancer patients) for whom SE-iFISH CTC and MassARRAY detection was simultaneously conducted, positivity rates were as follows: CTCs 78.57%, XPC A2815C 42.86%, and XRCC1 G1196A 7.12%. After these three biomarkers were combined, the positivity rate reached 92.86%, greatly improving diagnostic efficiency (Figure 2B, Table 4).
Table 4
| Variable | No. of participants | CTC patients | XPC A2815C patients | XRCC1 G1196A patients | ||
|---|---|---|---|---|---|---|
| Men | Women | All | ||||
| Bladder cancer patients | 5 | 3 | 8 | 6 | 3 | 0 |
| Renal carcinoma patients | 3 | 1 | 4 | 3 | 1 | 1 |
| Prostate cancer patients | 2 | 0 | 2 | 2 | 2 | 0 |
CTC, circulating tumor cell; SNP, single-nucleotide polymorphism.
SE-iFISH CTC displayed a valid effect on recurrence monitoring for genitourinary system cancer patients
Genitourinary system cancers have a high recurrence rate at 6 months to 2 years after surgery, and effective recurrence monitoring indicators are very important for screening cancer patients. We continued to explore the ability of SE-iFISH CTC to monitor genitourinary system cancers. Among detecting 12 bladder cancer patients with postoperative recurrence, 10 CTC-positive cases (66.67% positivity rate) and 5 urine cytology-positive cases (41.67% positivity rate) were observed (Figure 3A, Table 2).
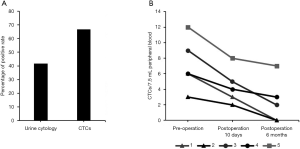
We then analyzed 5 patients by SE-iFISH CTC to assess their cancer progression at presurgery, 10 days postsurgery, and 6 months postsurgery stages. Patient 1 and patient 2, who experienced early prostatic cancer and showed CTCs preoperatively, CTCs were not detected after 6 months postoperatively, resulting in a good prognosis. In contrast, patient 5 had terminal renal carcinoma and more CTCs with a poor prognosis (Figure 3B). CTCs were detected in patients 3 and 4, who had cancer progression 6 months postsurgery. Therefore, the number of CTCs was consistently accurate and consistent with cancer progression and may be used in cancer monitoring.
Discussion
Many CTC detection methods are currently used, such as the CellSearch system, microfluidic isolation, and SE-iFISH. The CellSearch system has showed a low detection rate and sensitivity, and only 38% of CTCs could be detected (24), which is not suitable for CTC testing. Another method, microfluidic isolation, is based on specific antibody expression by CTCs or CTC size (28), however, not every CTC cell expresses tumor marker. SE-iFISH is based on subtractive enrichment, which can enrich more CTCs and shows excellent results (25). The positivity rate for genitourinary system cancer patients in the preliminary experiment was 70%, and that for healthy individuals was 0 (Figure 1B). When a large number of samples were analyzed, CTCs also exhibited a high detection rate (Table 2), and the number of CTCs was directly related to cancer progression (Figure 1C). Unfortunately, we only examined T1, T2, and T3 tumor stage patients. No T4 stage patients were enrolled; therefore, more patients need to be recruited for a more accurate study.
To increase screening sensitivity and without increasing trauma to patients, the MassARRAY iPLEX Gold multiple genotyping analysis system was used, and many cancer susceptibility genes were selected in urine cells (data not shown). Ultimately, XPC A2815C and XRCC1 G1196A were screened out and were able to effectively distinguish cancer patients from those without noncancer (Figure 2A). After combining analysis of XPC A2815C, XRCC1 G1196A, and CTCs, the positivity rate reached 92.86%, and these panels demonstrated excellent screening effectiveness for genitourinary system cancers (Figure 2B). In the course of the study, many cancer-sensitive genes, such as EGFR, ERBB2, TERT, or BRAF, were also selected as candidates; however, they were not used in this study due to their difficult detection or low specificity (mutations were detected in noncancer subjects) (data not shown). Of course, this result cannot exclude their application value and requires further exploration.
CTCs are effective tumor screening and tumor monitoring biomarkers for genitourinary system cancers. Although few clinical case descriptions exist in the literature, validations has been performed in other studies (10,11).
In conclusion, our study revealed that CTCs, which are excellent screening biomarkers, detected by SE-iFISH can be used in genitourinary system cancer screening and monitoring. Based on the MassARRAY system of urine cell detection, noninvasive XPC A2815C and XRCC1 G1196A SNP detection was used as the specific supplement for CTC detection.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.47). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committees of Quanzhou First Hospital Affiliated to Fujian Medical University and Xiangyang No. 1 People’s Hospital. All patients and healthy individuals signed informed consent before the experiment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Australian Medical Journal 1869;14:146-7.
- Wang C, Mu Z, Chervoneva I, et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res Treat 2017;161:83-94. [Crossref] [PubMed]
- Klameth L, Rath B, Hochmaier M, et al. Small cell lung cancer: model of circulating tumor cell tumorospheres in chemoresistance. Sci Rep 2017;7:5337. [Crossref] [PubMed]
- Msaouel P, Koutsilieris M. Diagnostic value of circulating tumor cell detection in bladder and urothelial cancer: systematic review and meta-analysis. BMC Cancer 2011;11:336. [Crossref] [PubMed]
- Chen JF, Lu YT, Cheng S, et al. Circulating tumor cells in prostate cancer: beyond enumeration. Clin Adv Hematol Oncol 2017;15:63-73. [Crossref] [PubMed]
- Soave A, Riethdorf S, Dahlem R, et al. Commentary on "Detection and oncological effect of circulating tumor cells in patients with variant urothelial carcinoma histology treated with radical cystectomy.". Urol Oncol 2018;36:347-8. [Crossref] [PubMed]
- Abrahamsson J, Aaltonen K, Engilbertsson H, et al. Circulating tumor cells in patients with advanced urothelial carcinoma of the bladder: Association with tumor stage, lymph node metastases, FDG-PET findings, and survival. Urol Oncol 2017;35:606.e9-606.e16. [Crossref] [PubMed]
- Nagaya N, Kanayama M, Nagata M, et al. The Surge in the Number of Circulating Tumor Cells Following Treatment with Sunitinib for Metastatic Renal Cell Carcinoma. Intern Med 2018;57:2695-700. [Crossref] [PubMed]
- Chen J, Cao S, Situ B, et al. Metabolic reprogramming-based characterization of circulating tumor cells in prostate cancer. J Exp Clin Cancer Res 2018;37:127. [Crossref] [PubMed]
- Heller G, McCormack R, Kheoh T, et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J Clin Oncol 2018;36:572-80. [Crossref] [PubMed]
- Costa WH, Jabboure GN, Cunha IW. Urological cancer related to familial syndromes. Int Braz J Urol 2017;43:192-201. [Crossref] [PubMed]
- Geary J, Sasieni P, Houlston R, et al. Gene-related cancer spectrum in families with hereditary non-polyposis colorectal cancer (HNPCC). Fam Cancer 2008;7:163-72. [Crossref] [PubMed]
- Maia MC, Bergerot PG, Dizman N, et al. Association of Circulating Tumor DNA (ctDNA) Detection in Metastatic Renal Cell Carcinoma (mRCC) with Tumor Burden. Kidney Cancer 2017;1:65-70. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Kim CJ, Park J, Sunkara V, et al. Fully automated, on-site isolation of cfDNA from whole blood for cancer therapy monitoring. Lab Chip 2018;18:1320-9. [Crossref] [PubMed]
- Eeles R, Goh C, Castro E, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol 2014;11:18-31. [Crossref] [PubMed]
- Li Q, Ma R, Zhang M. XRCC1 rs1799782 (C194T) polymorphism correlated with tumor metastasis and molecular subtypes in breast cancer. Onco Targets Ther 2018;11:8435-44. [Crossref] [PubMed]
- Sanjari Moghaddam A, Nazarzadeh M, Sanjari Moghaddam H, et al. XRCC1 Gene Polymorphisms and Breast Cancer Risk: A Systematic Review and Meta- Analysis Study. Asian Pac J Cancer Prev 2016;17:323-30. [Crossref] [PubMed]
- Yoshino Y, Takeuchi S, Katoh T, et al. XPC intron11 C/A polymorphism as a risk factor for prostate cancer. Environ Health Prev Med 2016;21:100-4. [Crossref] [PubMed]
- Kahnamouei SA, Narouie B, Sotoudeh M, et al. Association of XPC Gene Polymorphisms with Prostate Cancer Risk. Clin Lab 2016;62:1009-15. [Crossref] [PubMed]
- Andree KC, van Dalum G, Terstappen LW. Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol 2016;10:395-407. [Crossref] [PubMed]
- Sheng Y, Wang T, Li H, et al. Comparison of analytic performances of Cellsearch and iFISH approach in detecting circulating tumor cells. Oncotarget 2017;8:8801-6. [Crossref] [PubMed]
- Ye Z, Ding Y, Chen Z, et al. Detecting and phenotyping of aneuploid circulating tumor cells in patients with various malignancies. Cancer Biol Ther 2019;20:546-51. [Crossref] [PubMed]
- Descotes F, Kara N, Decaussin-Petrucci M, et al. Non-invasive prediction of recurrence in bladder cancer by detecting somatic TERT promoter mutations in urine. Br J Cancer 2017;117:583-7. [Crossref] [PubMed]
- Min KW, Kim WS, Jang SJ, et al. MassARRAY, pyrosequencing, and PNA clamping for EGFR mutation detection in lung cancer tissue and cytological samples: a multicenter study. J Cancer Res Clin Oncol 2016;142:2209-16. [Crossref] [PubMed]
- Jiang X, Wong KHK, Khankhel AH, et al. Microfluidic isolation of platelet-covered circulating tumor cells. Lab Chip 2017;17:3498-503. [Crossref] [PubMed]


