
STAT3 activation regulated circ-STAT3.46 promote expression of IGF1R by sponging of miR-139-5p in human colon cancer
Introduction
Decades ago, Nigro et al. first discovered a new type of circular non-coding RNAs (circRNAs) which were regarded as spliced transcripts of a candidate gene (1). However, most of the researches considered circRNAs as junk RNA due to its characters of Non-function. After decades of further study and the advances in high-through sequencing, circRNAs were regarded as a vital part of non-coding RNAs which specifically expressed in specific tissues (2,3). circRNAs were more stable than the linear mRNAs for their resistance to RNase activity, which distributed mainly in the cytoplasm, but some seemed to be enriched in the nucleus (2).
Clinical studies revealed that circRNAs were closely associated with human diseases, especially cancers including bladder carcinoma (4), esophageal squamous cell carcinoma (5), colorectal cancer (CRC) (6), hepatocellular carcinoma (7) and blood malignancies (8), etc. Moreover, these studies revealed that circRNAs were more abundant in the peripheral blood suggesting that they can serve as new biomarkers cancer diagnosis (9-11). Several circRNAs were discovered abnormally expressed in human colorectal cancer. Wang et al. found that hsa_circ_001988 was decreased in tumor tissues, suggesting that hsa_circ_001988 may be a potential treatment target and biomarker for colorectal cancer. The value of serum exosomes in colorectal cancer was evaluated, indicating that the serum exosome circRNAs level can distinguish CRC from healthy controls (12). The mechanistic study indicated both diagnosis value and mechanical mode of exosome circ-ITCH in human CRC. Downregulation of circ-ITCH in human CRC can inhibit the Wnt/β-catenin pathway by up-regulating ITCH (13).
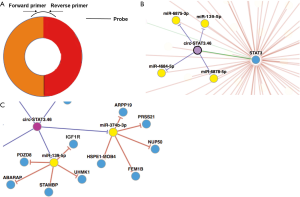
So far, the functions of CircRNAs can be concluded as below: (I) miRNA sponges, (II) RNA-binding protein (RBP) sponges, (III) regulators of transcription, and (IV) proteins/peptides translation potent (2,3). Among these four functions, miRNA sponge is mostly addressed in previous studies. Using a circRNAs database, circNet (14), we obtained that the regulatory network consisted of circ-STAT3.46-miR-139-5p-IGFR1. Predictively, circ-STAT3.46 have the potential to sponge a bunch of miRNAs including miR-139-5p, miR-374-3p, miR-3617, miR-619-3p and miR-3136-3p. By preliminary screening, we found that only miR-139-5p was significantly correlated with circ-STAT3.46 in human colon cancer. Furthermore, we found that the receptor of IGF1, IGF1R, was overexpressed in human colon cancer. Therefore, in the present study, the potential circ-Stat3.46-miR-139-5p-IGF1R axis was investigated, we try to find out its diagnosis value and tumor promotion mechanism within human colon cancer.
Methods
Patients
The hospital-based case-control study has consisted of 56 colon cancer patients and 56 cancer-free controls. All the subjects were recruited from the Zhenjiang Hospital of Traditional Chinese Medicine between January 2010 and September 2016. All the patients were enduring surgery treatment for primary colon cancer. This study was approved by the Ethics Review Board of Zhenjiang Hospital of Traditional Chinese Medicine (2017-008), and every patient had written informed consent.
Potential circRNA-miRNA-gene regulatory prediction
The possible interaction between circ-STAT3.46 and sponge miRNA was predicted by using an online prediction tool, CircNet (http://syslab5.nchu.edu.tw/CircNet/). The information of circRNA was obtained in circRNA (http://www.circrna.org).
Cell lines and reagents
Human CC cell lines including Hct-116 and Hct-15 were purchased from American Type Culture Collection (ATCC). All cells were maintained in Dulbecco modified Eagle medium purchased from Gibco (CA, USA) supplemented with 10% fetal bovine serum (Thermofisher, CA) and maintained in humidified 5% CO2 at 37 °C. Human Recombinant IGF1 was purchased from Abcam (ab9573, MA), and inhibitor for JAK2/STAT3 signaling AZD1480 was purchased from Selleckchem company (Cat. No. S2162).
Serum exosome extraction
The experiment was carried out strictly following the instructions of Total Exosome Isolation Reagent (from serum) (4478360 Invitrogen, CA). One mL of serum sample was mixed with 200 uL exosome isolation reagent, incubate at 4 for 30 min, centrifuge at 10,000 g for 10 min at room temperature, the pallet was dissolved by Trizol for further analysis.
Real-time PCR
The total RNA was isolated with TRIzol reagent. And the expression of circ-STAT3.46 was detected by using Sybergreen® based real-time PCR. The primers involved in the study were: forward primer: gagactgtaccactgca, reverse primer: ggccaacatggtgaaacgc, amplicon size 127bp for circ-STAT3.46, the primer design for this circ-RNA was presented in supplementary Figure 1A, for detection of internal control gene GAPDH: forward Primer TGTGGGCATCAATGGATTTGG, reverse Primer ACACCATGTATTCCGGGTCAAT, amplicon size 110 bp. The transcription of miR-139-5p was detected by using a commercial Taqman probe (4427975) (ThermoFisher, CA). The miR-139-5p-3p mimic was also purchased from Thermo Fischer (MC11749).
Western blot
Cells treated variously were collected and lysed with cell lysis buffer (RIPA) (Thermofisher, CA), and further separated on 10% SDS-polyacrylamide gels and transferred on a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Immunoblotting was performed using the following primary antibodies: anti-IGF1R (#9750, Cell signaling, MA).
RNA pull-down
The RNA pull-down assay was performed by using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermofisher, CA) according to the instructions from the manufacturer. Briefly, the total RNA was extracted from HCT-116 cells, after the treatment of RNase in room temperature, magnetic beads were incubated with Probes for biotin-labeled circ-STAT3.46 (Genescript Co., Nanjing, China) (sequence: ccagactccatctcggccaggcatgg) and U6 control (sequence: ttggaacgatacagagaagatt). The relative expression levels of miR-139-5p, circ-STAT3.46 and U6 in the extract were detected by RT-PCR.
Immunohistochemistry staining
Tissue sections were following a deparaffinized and rehydrated process, and then block endogenous peroxidase activity by being treated with 3% H2O2 in methanol for 30 min to. Tissue sections were then rinsed twice for five minutes in phosphate-buffered saline (PBS) and blocked with 10% normal goat serum. After resin, the samples were incubated with a primary anti-STAT3 (Y705) (#9131, Cell signaling, MA) at 4 °C overnight. Sections were incubated with secondary antibodies and stained with DAB according to the manufacturer’s protocol, mounted on slides, and photographed using a digital microscope camera (Nikon, Tokyo, Japan).
Luciferase reporter assay and transfection of miR-139-5p
The 3'UTR region of IGF1R containing the wild-type or mutant potential target site for miR-139-5p were designed and synthesized in Genescript Co. (Nanjing, China) and inserted into the a pGL4 vector (Promega, WI). For luciferase assay, Hct-116 and Hct-15 were co-transfected with pGL4-IGF1R-WT 3'UTR or pGL4-IGF1R-MU 3'UTR, with miR-139-5p mimics or control (GenePharma, China) using Lipofectamine 2000 (Thermofisher, CA). Cells were harvested 48 h after transfection for luciferase assay using a Dual-Luciferase® Reporter Assay System (Promega, WI) according to the manufacturer’s protocol.
Statistical analysis
Data are presented as mean ± SD. The statistical differences in demographic and clinical characteristics were evaluated by using χ2 tests and the Student’s t-test analysis. The two values correlations were assessed by linear correlation analysis and followed by F-test. The overall survival rates in different groups were analyzed by using Kaplan-Meier curve. All the expression experiments we conducted in vitro were repeated at least three times with samples in triplicates. Statistical analysis was performed using the GraphPad Prism software. In all cases, P<0.05 was considered statistically significant.
Results
Characterization of circ-STAT3.46 in human colon cancer
We found that has-circ-STAT3.46 is located in 17q21.2, the position is chr17:40498586-40500557, it is also nominated as hsa_circ_0043817. After getting its full length (296 bp) the primers detecting its expression and the probes for RNA pull-down assay were designed (Figure S1A). The regulatory potential of circ-STAT3.46 was predicted by using the on-line tool, CircNet. We found that circ-STAT3.46 has the potential to sponge a serial of miRNAs including miR-139-5p, miR-374-3p, miR-3617, miR-619-3p and miR-3136-3p (Figure S1B). After screening, we found that only miR-139-5p was decreased in human colon cancer tissues, while others were not. Moreover, we found that miR-139-5p have several potentially regulatory targets including PDZD8, ABARAP, STAMBP, UHMK, and IGFR1 (Figure S1C). Since several studies have already reported that IGF1/IGF1R is a STAT3 activator, we proposed that overexpression of circ-STAT3.46 might be another explanation of STAT3 activation in human malignancies. Thus, we attended to pick circ-STAT3.46-miR-139-5p-IGF1R as a potential regulatory axis in human colon cancer for further investigation.
Overexpression of circ-STAT3.46 was associated with downregulation of miR-139-5p in human colon cancer tissues
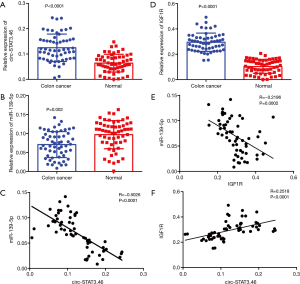
To investigate the relationship between circ-STAT3.46 and miR-139-5p, the transcription of both indices was detected by using real-time PCR; the results indicated that circ-STAT3.46 was overexpressed in human colon cancer tissues compared to the adjacent normal controls (Figure 1A). Meanwhile, miR-139-5p was significantly decreased in colon cancer compared to the adjacent normal controls (Figure 1B). Moreover, negatively correlation was found between the expression of the circ-STAT3.46 and miR-139-5p within human colon cancer tissues (r=−0.5026, P<0.0001) (Figure 1C). Meanwhile, the transcription of IGF1R was also determined, we found that IGF1R was not only overexpressed (Figure 1D), but also positively correlated to circ-STAT3.46 expression (Figure 1E) and negatively correlated to miR-139-5p expression (Figure 1F) in human colon cancer tissues.
Serum Circ-STAT3.46 was associated with metastasis and served as a poor prognosis of colon cancer patients
Since circ-STAT3.46 was overexpressed in colon cancer tissues, we further analyzed the relation between circ-STAT3.46 expression and clinical characters of colon cancer patients. Figure 2A showed that circ-STAT3.46 was significantly higher in the colon cancer tissues of TMN stage III-IV compared to I-II. We also studied the relation between circ-STAT3.46 and the prognosis of the post-surgery patients. The result indicated that higher expression of circ-STAT3.46 was associated with poor prognosis of the post-surgery colon cancer patients (P=0.037, HR =2.272, 95% CI: 1.052 to 4.839) (Figure 2B).
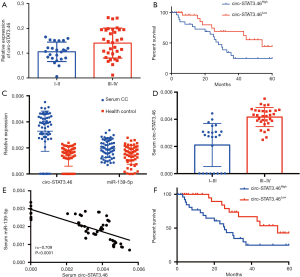
Since circRNA and miRNA were reported to stably exist in human serum, the serum RNA was extracted, we found that existence of circSTAT3.46 and miR-139-5p was similar to the colon cancer tissues, overexpression of circSTAT3.46 and downregulation of miR-139-5p were found in the serum of colon cancer patients compared to the normal controls (n=56 for each group) (Figure 2C). Also, the serum circSTAT3.46 was significantly higher in the TMN III-IV patients compared to the patients of I-II stage (Figure 2D). And the level of serum circ-STAT3.46 was negatively correlated to serum miR-139-5p level in human colon cancer patients (Figure 2E). We also found that serum circSTAT3.46 was a valuable indicator in predicting the prognosis of the post-surgery colon cancer patients (Figure 2F) (P=0.004, HR =2.272, 95% CI: 1.261 to 5.646).
Overexpression of circ-STAT3.46 was related to IGF1 dependent STAT3 activation in human colon cancer
Because circ-STAT3.46 are predicted to regulate IGF1R, and IGF1/IGF1R were reported to be an activator for STAT3 activation (Y705), we presumed that circ-STAT3 might relate to STAT3 expression or activation. Therefore, the associations between STAT3 expression or activation and the expression of circr-STAT3.46 were studied. The results indicated that STAT3 activation (STAT3 nuclear localization) was significantly stronger in circ-STAT3.46High group compared to the circ-STAT3.46low group (Figure 3A), but there was no difference in STAT3 expression between these two groups (data not shown). Positive correlation was also spotted between circ-STAT3.46 transcription and STAT3 activation (nuclear positive percentage) in human colon cancer tissues (r=0.605, P<0.0001) (Figure 3B).
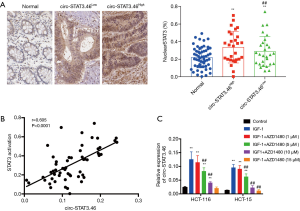
Furthermore, the in vitro study for IGF-1/IGF1R/STAT3 regulated circ-STAT3.46 transcription was carried out. First, the treatment of IGF-1 (100 ng/mL) can significantly increase the transcription of circ-STAT3.46 in both HCT-116 and HCT-15 cells, but the transcription can be blocked with the treatment of JAK2/STAT3 inhibitor, AZD1480, especially when the concentration increased to 5 µM (Figure 3C).
circSTAT3.64 downregulate miR-139-5p and upregulate IGF1R in vitro study
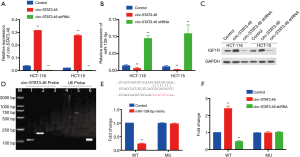
Since we have verified the potential regulatory axis, circ-Stat3.46-miR139-5p-IGF1R from the investigation of clinical samples, to confirm such regulatory axis in vitro, first, we investigated the relationship between circ-STAT3.46 and miR-139-5p in human colon cancer cell line: Hct-116 and Hct-115. miR-139-5p decreased significantly in HCT-116 and HCT-15 circ-STAT3.46 overexpression cells (HCT-116-circ-Stat3.46 and HCT-15-circ-STAT3.46) and increased significantly when shRNA of circ-STAT3.46 was involved (HCT-116-circ-STAT3.46-shRNA and HCT-15-circ-STAT3.46-shRNA) (Figure 4A,B). Also, the protein expression of IGF1R was detected by western-blot, increased IGF1R was detected in the circ-STAT3.46 overexpression colon cell lines, and its expression decreased once circ-STAT3.46 was knock-down (Figure 4C). Next, the direct binding between circ-STAT3.46 and miR-139-5p was verified in HCT-116-circ-STAT3.46 cells by using biotinylated RNA Pull-Down Assay, and miR-139-5p can be detected among the precipitated RNAs using RT-PCR (Figure 4D). To verify the regulation of miR-139-5p on IGF1R, the binding potent of miR-139-5p in IGF1R 3'UTR was predicted by using TARGETSCAN. The mutation of IGF1R 3'UTR was generated based on the prediction, a significant decrease of IGF1R promoter activity was found in WT and was fully restored in the mutated form (MU) (Figure 4E), this result indicated that miR-139-5p can downregulate IGF1R by binding to its 3'UTR. Moreover, a luciferase reporter assay was performed using pGL4-Luc plasmid containing WT and miR-139-5p binding site mutated IGF1R 3'UTR (pGL4-IGF1R 3'UTR-WT, WT and pGL4-IGF1R 3'UTR-MU, MU) in various cells, the activity of WT increased significantly in HCT-116-circ- STAT3.46 and HCT-15-circ-STAT3.46 cells, and decreased significantly in HCT-116-circ-STAT3.64-shRNA and HCT-15-circ-STAT3.46-shRNA cells. But there is no significant change in promoter activity in pGL4-IGF1R 3'UTR-MU transfected cells (Figure 4F).
In summary, the axis of circ-Stat3.46-miR-139-5p-IGF1R was existed in human colon cancer, and STAT3 regulated circ-STAT3.46 might enhance the expression of IGF1R by sponging miR-139-5p.
Discussion
Insulin-like growth factor 1 (IGF-1) is a hormone which structurally similar to insulin (15). Physiologically, it played an essential role in body growth which primarily produced in the liver (15). Pathologically, increased expression of IGF-1 was discovered in many human malignancies, IGF1R is the receptor of IGF1, IGF1/IGF1R mediated signaling controls many biological processes of tumor, such as proliferation and metastasis (15). These effects were regulated by its down-stream signaling including wnt/β-Catenin, PI3K/AKT, MAPK, and STAT3 signaling (16,17). In the present study, we found that circ-STAT3.46 transcription can be enhanced by IGF1/IGF1R/STAT3 signaling, specifically inhibit JAK2/STAT3(Y705) activation can effectively block the transcription of circ-STAT3.46. We suspected that high expression of circ-STAT3.46 in colon cancer was a protective mechanism which can guarantee the IGF1/IGF1R dependent STAT3 activation. Our results indicated that circ-STAT3.46 could bind to miR-139-5p, thus to maintain the high expression level of IGF1R, which can response for the exogenous IGF-1 stimulation.
miR-139-5p was usually reported as a tumor suppressor which was first characterized in neurodegeneration (18). Subsequently, miR-139-5p was identified to be responsible for cancer diagnosis, prognosis, and therapy (19). Several targets of miR-139-5p have been discovered, its characteristic is contradictory, due to its regulation on sets of cell proliferation and apoptosis-related genes (20). One of these researches revealed that miR-139-5p inhibited cell proliferation and invasion by targeting IGF1R in human non-small cell lung cancer (21). Maoa et al. reported that in miR-139-5p inhibited the crosstalk between PI3K/AKT and Wnt pathway mediated by IGF-1R in HCT-116 cells (22). In our study, we provided both clinical and experimental proofs for the miR-139-5p regulation on IGF1R. And furthermore, we discovered one of the reasons of downregulation of miR-139-5p. The overexpression of circ-STAT3.46 can sponge miR-139-5p by direct binding.
We also explored the value of serum circ-STAT3.46 and miR-139-5p as biomarkers in prediction different TMN stages and prognosis of colon cancer patients. As it has been introduced previously, compared to common linear mRNAs, miRNA and circRNA are steadier and more RNase resistant (2,3). Many researches indicated that they were secreted and detected in peripheral blood. Based on the detection of serum circ-STAT3.46 and miR-139-5p, we can quickly evaluate the expression level of IGF1R as well as the activation of STAT3 signaling within the colon cancer tissues. Many other groups have developed circulating miRNA diagnosis system for many human malignancies, including colon cancer, hepatocellular carcinoma, Non-small cell lung cancer, prostate cancer, etc. (9-11). Concerning circRNA, the use of circRNA as biomarkers has emerged to be one of the main approaches for diagnosis and prognosis of cancers. circRNA meet the essential features to be a qualified biomarker such as stability, sensitivity, specificity, accuracy and reproducibility. Tang et al. found that circ-KIAA1244 was lower expressed in human gastric cancer tissues, plasmas, and the result also demonstrated that a decreased expression of circ-KIAA1244 in plasmas exosome was negatively correlated with TNM stage and metastasis, and a bad prognosis of GC patients (23). The report concerning the roles of circulating circRNA in human colon cancer is relatively rare, therefore, we reported the diagnosis value of circulating circ-STAT3.46 in human colon cancer for the very first time.
Conclusively, we explored an axis consisted of circ-Stat3.46-miR-139-5p-IGF1R, which was closely related to cell proliferation initiated by STAT3 activation. Moreover, the serum circ-STAT3.46 and miR-139-5p can serve as potential biomarkers for colon cancer diagnosis.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.31). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Review Board of Zhenjiang Hospital of Traditional Chinese Medicine (2017-008), and every patient had written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol 2017;14:1000-6. [Crossref] [PubMed]
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679-92. [Crossref] [PubMed]
- Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017;403:305-17. [Crossref] [PubMed]
- Wang W, Ma J, Lu J, et al. Circ0043898 acts as a tumor inhibitor and performs regulatory effect on the inhibition of esophageal carcinoma. Cancer Biol Ther 2018;19:1117-27. [Crossref] [PubMed]
- Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun 2018;505:346-52. [Crossref] [PubMed]
- Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol 2018;234:500-8. [Crossref] [PubMed]
- Zhang X, Zhou H, Jing W, et al. The Circular RNA hsa_circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and May Serve as a Diagnostic Biomarker. Dis Markers 2018;2018:3073467.
- Chen D, Zhang C, Lin J, et al. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res 2018;10:1275-83. [Crossref] [PubMed]
- Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 2017;466:167-71. [Crossref] [PubMed]
- Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 2018;18:116. [Crossref] [PubMed]
- Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 2015;8:16020-5. [PubMed]
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 2018;17:19. [Crossref] [PubMed]
- Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016;44:D209-15. [Crossref] [PubMed]
- Kineman RD, Del Rio-Moreno M, Sarmento-Cabral A. 40 YEARS of IGF1: Understanding the tissue-specific roles of IGF1/IGF1R in regulating metabolism using the Cre/loxP system. J Mol Endocrinol 2018;61:T187-98. [Crossref] [PubMed]
- Kumar AS, Rayala SK, Venkatraman G. Targeting IGF1R pathway in cancer with microRNAs: How close are we? RNA Biol 2018;15:320-6. [Crossref] [PubMed]
- Vishwamitra D, George SK, Shi P, et al. Type I insulin-like growth factor receptor signaling in hematological malignancies. Oncotarget 2017;8:1814-44. [Crossref] [PubMed]
- Saba R, Goodman CD, Huzarewich RL, et al. A miRNA signature of prion induced neurodegeneration. PLoS One 2008;3:e3652. [Crossref] [PubMed]
- Huang LL, Huang LW, Wang L, et al. Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncol Lett 2017;14:1215-22. [Crossref] [PubMed]
- Zhang HD, Jiang LH, Sun DW, et al. MiR-139-5p: promising biomarker for cancer. Tumour Biol 2015;36:1355-65. [Crossref] [PubMed]
- Xu W, Hang M, Yuan CY, et al. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:3864-70. [PubMed]
- Maoa R, Zou F, Yang L, et al. The loss of MiR-139-5p promotes colitis-associated tumorigenesis by mediating PI3K/AKT/Wnt signaling. Int J Biochem Cell Biol 2015;69:153-61. [Crossref] [PubMed]
- Tang W, Fu K, Sun H, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer 2018;17:137. [Crossref] [PubMed]


