
Overexpression of hyaluronan-binding protein 1 promotes the proliferation, migration, and invasion of malignant glioma
Introduction
Glioma is the most common intracranial malignancy, which originates from neuroepithelial tumors—the annual incidence of glioma in 3–8 persons/100,000 (1). Like most tumors, gliomas are caused by the interaction of innate genetic factors and environmental factors. Because of the different degrees of malignancy, the symptoms and conditions of the patients are different. Surgery is the first step in the treatment of most gliomas. Some low-grade gliomas can be treated by surgery. However, grade IV gliomas account for 50% of all malignant gliomas (2), while grade II–IV gliomas are prone to recurrence (3). Extended resection is a common method for many neurosurgical treatments of glioma. Even so, due to the characteristics of glioma infiltration and growth, enlarged resection cannot fundamentally prevent glioma growth (4-6), and further radiotherapy and chemotherapy are needed. Therefore, in-depth studies of the development of malignant glioma and related molecular mechanisms have become a key issue in the treatment of glioma.
Hyaluronic acid binding protein 1 (HABP1), a specific hyaluronic acid binding protein of the cell surface, is widely found in the brain, cartilage, blood vessels, and skin (7-9). HABP1 is the same as the splicing factor-related protein (p32) and complements the component globular head receptor (gc1qr) in tumorigenesis and metastasis (10,11). Given this, some scientists have named this protein P32 or gC1qR or HABP1 or HABP1/p32/gC1qR. HABP1 is the only glycosaminoglycan that is present in animal tissues and bacteria. It is localized to human chromosome 17p12–13 and is expressed on most cytoplasmic surfaces in free form or in non-covalent complexes (12). There is a specific structure of HABP1 containing two α-helices and two β-sheets, which has a large amount of negative charge, which is the key to the targeting of HABP1 and other protein tissues (13). HABP1 regulates a variety of cell biological properties such as cell movement, adhesion, migration, differentiation, and wound healing (14). It also plays an important regulatory role in the inflammatory response, tumor angiogenesis, and the invasion and metastasis of tumor cells. In addition, small molecule chemotherapy drugs have poor water solubility and low stability, while HABP1 has good water solubility and bioavailability. At present, HABP1 has received more and more attention in the field of cancer research. It has been proven that HABP1 appears in various tumors. Changes in characteristic expressions, such as increased expression of HABP1, promote growth and metastasis of breast cancer (15), endometrial cancer (16), and gastric cancer (17). Increase in HABP1 content is associated with melanoma hyperplasia and malignancy (18). We hypothesized that HABP1 might be the link between the extracellular matrix (ECM) and the cytoskeleton (19), and its molecular mechanism may be one of the important mechanisms for the development and prognosis of glioma.
Methods
Patients and tissue samples
All 80 cases of fresh frozen tissues came from the Affiliated Hospital of Nantong University. According to a protocol approved by the Human Research Committee of the Institutional Review Board (IRB), all human glioma tissues came from patients diagnosed between 2010 and 2018. All human glioma tissues came from patients who had not received radiotherapy or chemotherapy. Immunohistochemical staining sections were made from paraffin-embedded sections of fresh tissues after fixation. Histological types were determined according to the WHO review by two senior pathological directors. For the immunoblotting analysis of proteins, a 200 mg tissue block was taken from the tissue homogenizer, and the precooled homogenate medium was added to fully crush the tissues. The centrifuged homogenate was discarded and precipitated to leave the supernatant, which was frozen at −80 °C.
Immunohistochemistry
Firstly, we put the slices into an oven for 6–8 hours, and then put them into two xylene cylinders and soaked them for 15 minutes. After that, the slices were drained as much as possible and were put into 95%, 85%, and 75% alcohol cylinders. They were kept in each cylinder for 2 minutes, rinsed out after running, and placed in a container of distilled water. Secondly, the slices were placed in a 1X citrate buffer (pH 6.0). Using a high-pressure boiling method, the slices were heated for 5 minutes, and then naturally cooled. The cooled slices were removed and washed with PBS (pH 7.2). The remaining PBS was removed from the slices, and 100–200 µL of 3% H2O2 was added to each slice for 30 minutes. Then, they were washed for 3 times with PBS. Thirdly, the slices were put into a humidified chamber, and HABP1, Ki-67, and E-cadherin diluted antibodies were added to them. The humidified chamber was kept in a refrigerator at 4 °C overnight. The next day, the sections were removed, rewarmed for 30 minutes, washed with PBS, mixed with 100 µL of antibodies and 100 µL of horseradish peroxidase, incubated at room temperature for 20 minutes, washed with PBS, mixed with 100 µL of freshly prepared DAB for 1–2 minutes, and placed in a container with tap water. Fourthly, the slices were rinsed with running water, then counterstained with hematoxylin. The pieces were mixed with a proper amount of neutral gum and sealed to dry. Finally, the staining was observed under a microscope. The cell scoring ratios were as follows: 0 (<1%), 1 (1–25%), 2 (26–50%), 3 (51–75%) and 4 (>75%). Dyeing intensity was graded according to the following criteria: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellowish brown), and 3 (strong staining = brown).
Western blot analysis
Homogenized protein was added to the separation gel. Then the separation gel was installed on the electrophoresis apparatus. Prepared electrophoresis solution (10% sodium dodecyl sulfate, glycine, and tris) and transfer buffer (methanol, glycine, and tris) were added to the electrophoresis apparatus box. The homogenized protein was transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were put into 5% skim milk for 2 hours. Then, primary antibodies were added to the membranes, and the membranes were kept at 4 °C overnight. The next day, the membranes were washed with PBST. The secondary antibodies were added to the membranes. The membranes were incubated at room temperature for 2 hours. The experiment was replicated independently 3 times, and the results were analyzed by a computer-aided image analysis system (Adobe Systems, SanJose, CA, USA).
Antibodies
The antibodies used in the study included anti-HABP1 antibody (1:500; Santa Cruz Biotechnology), E-cadherin (1:500; Santa Cruz Biotechnology), GAPDH (1:2,000; Santa Cruz Biotechnology), PCNA (1:1,000; Santa Cruz Biotechnology), Cyclin D1 (1:1,000; Santa Cruz Biotechnology), and MMP-9 (1:500, Santa Cruz Biotechnology).
Cell culture
The glioma cell lines H4, U251MG, U87MG, A172, and HEK293T were purchased from the Cell Bank of the Chinese Academy of Sciences. All cells were cultured in DMEM (Life Technologies) containing 10% fetal bovine serum (FBS) in a 37 °C 5% CO2 incubator. HEK293T cells were cultured in RPMI 1640 with 10% FBS. The cells were digested with 0.25% trypsin, and the medium was changed every 2–3 days.
Expression plasmid and transient transfection
All three oligonucleotides containing the short hairpin RNA (shrna) target sequence were designed by GeneChem (Shanghai, China). The sequences were as follows: siHABP1-1, CCCAACTTTGTGGTTGAAGTT; siHABP1-2, GCTAAATTATTGCGCAAAGTT; siHABP1-3, GCACCAGGAATATATCACCTT. The cells were transfected with Lipofectamine 2000 (Life Technologies). The transfection lasted for 48 hours as recommended by the manufacturer.
Cell proliferation assay
The cells were digested and put into 96-well cell plates (Corning Inc., Corning, NY, USA), cultured overnight. The next day, the Cell Counting Kit CCK-8 (Dojindo, Kumamoto, Japan) was added to the cells. A 37 °C incubator was used to incubate the cells for 2 hours in the dark, and then a 450 nm automatic plate reader was used to read the data.
Transwell migration assay
We digested glioma cells, counted them, made a cell suspension, added 100 µL of cell suspension per well, added 500 µL of medium containing 10% fetal bovine serum to the lower chamber, and incubated them in a 37 °C incubator for 24 hours. Next, the cells were washed 3 times with PBS, fixed with 5% glutaraldehyde for 1 hour, and stained with 0.1% crystal violet for half an hour. Finally, cotton swabs were used to wipe off the surface cells, and the cells were observed under a microscope. Three independent experiments were replicated.
Monolayer wound-healing assays
Cells were added to the six-well plate. The next day, a sterile probe was used to scribe the cells perpendicular to the cell plane. Next, the cells were washed with sterile PBS 3 times, cultured in a 37 °C 5% CO2 incubator, and removed 24 hours later. The microscope line observed and measured the width of the scratches and took a picture. Three experiments were repeated independently.
Immunofluorescence staining
The cells were digested and put into a 24-well plate. We then placed a sterile disc in each well. The cells were cultured overnight. The next day, the cells were washed with pre-cooled PBS. The discs were fixed in 4% paraformaldehyde for 1 hour, and permeabilized for 15 minutes with 0.1% Triton. Next, the discs were taken out, mixed with primary antibodies, and incubated at 4°C overnight. The next day, the discs were washed with PBS 3 times. The secondary antibodies and Hoechst 33342 dye were added to the discs and the discs were kept in the dark for 2 hours at room temperature. A Leica microscope (Germany) was used to observe and photograph the results.
Statistical analysis
All data were analyzed by SPSS13.0 (SPSS Inc., Chicago, IL, USA), and the chi-square test was used to evaluate the differences in clinicopathological features of HABP1. Survival analysis was performed using the Kaplan-Meier method, and the differences were compared using a log-rank test. P values <0.05 were considered as statistically significant.
Results
Expression of HABP1 in human glioma tissues
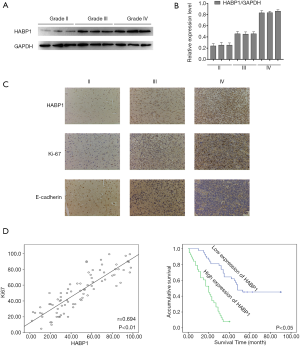
First, we investigated the expression of HABP1 in human glioma tissues at different levels by Western blot analysis. The results showed that the higher the glioma grade, the stronger the expression of HABP1 (Figure 1A,B). We speculated that HABP1 promotes glioma development. Then, we selected grades II, III, and IV to study the expression of HABP1 and Ki-67. Immunohistochemical staining showed that the immunohistochemical staining of grade IV was significantly stronger than that of grade II (Figure 1C). The expression of HABP1 was negatively correlated with E-cadherin, but positively correlated with Ki-67 (Figure 1D), a commonly used indicator of tumor proliferation. We then evaluated the clinical data of 80 patients with glioma and the expression of HABP1 by SPSS analysis. The clinicopathological features of HABP1 were evaluated (Table S1). Of the 80 glioma patients, 42 cases had low HABP1 expression and 38 cases had high HABP1 expression. High HABP1 expression was not significantly associated with patient age, gender, resection range, tumor size, or location. However, HABP1 expression was significantly associated with WHO stage (P=0.034). Cox’s proportional hazard regression model (Table S2) showed that WHO classification was an independent prognostic factor affecting the survival rate of glioma patients. The Kaplan-Meier survival curves indicated that the expression of low HABP1 was consistent with the prolonged survival of patients (Figure 1E). These data suggest that HABP1 may mediate the development of glioma.
Expression of HABP1 in different glioma cell lines
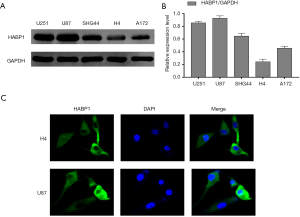
In order to further explore the effects of HABP1 on glioma, we chose to culture 5 different glioma cell lines including U87MG, U215MG, SHG44, H4, and A172, and analyzed the protein expression of HABP1 by Western blotting. The results showed that the U87 cell line had high protein expression while the H4 cell line had low expression of HABP1 protein (Figure 2A,B). We then used H4 and U87MG cell lines for immunofluorescence staining to visualize the morphological expression of HABP1 on glioma cell lines (Figure 2C).
Knockdown of the expression of HABP1 inhibits glioma cells
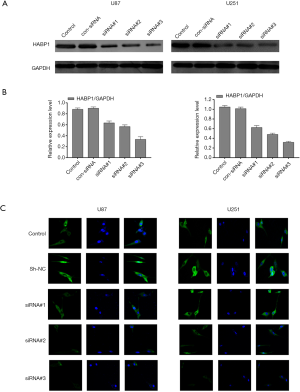
To further demonstrate the proliferative effect of HABP1 on glioma cells, we used short interfering RNA to knock down HABP1 and used the level of protein expression to investigate the interference of small interfering HABP1 on glioma cell lines. First, we transfected three silencing sites of HABP1 into U87MG and U251MG glioma cells with a high expression of HABP1. The transfection time was 48 hours, and a control group, an empty plasmid group, and an interference group were set up (Figure 3A,B). The results showed that the expression of HABP1 on glioma decreased after 3 groups of interference, and the effect of the siRNA-3 HABP1 interference plasmid was stronger than that of siRNA-1 and siRNA-2. Next, we transfected the U251MG and U87MG cell lines with three silencing HABP1, performed immunofluorescence staining, and visually observed the transfection efficiency of small interfering HABP1 on glioma cell line (Figure 3C). The experiment was repeated 3 times.
HABP1 knockdown inhibits proliferation, invasion, and migration of U87MG glioma cells
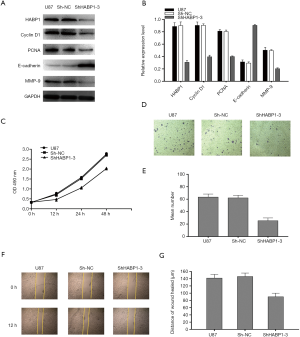
To further explore the mechanism of action of HABP1 on the development of glioma, we selected the small interfering HABP1 to transfect U87MG cells with a high expression of endogenous HABP1, collected cellular proteins, and analyzed the expression of PCNA, CyclinD1, MMP-9 by Western blot. It was found that MMP-9, PCNA, CyclinD1 expression decreased, and E-cadherin expression increased (Figure 4A,B). CCK-8 assay transfected U87MG cells, and cell viability was detected at 0 hours, 12 hours, 24 hours, 48 hours; the results indicated that silencing HABP1 potently inhibited cell viability (Figure 4C). In order to further explain the regulation of HABP1 migration in glioma cells, we used transwell assay to determine the 24-hour cell migration number of the control group, the empty plasmid group, and the interference group. The results showed that the cell migration ability of the interference group was significantly lower than that of the other two groups (Figure 4D,E). We then used the same transfection method to observe the cell invasion ability from the perspective of cell biology. After 24 hours, the wound healing ability of U87MG cells with small interference HABP1 was significantly lower than that of the control group (Figure 4F,G).
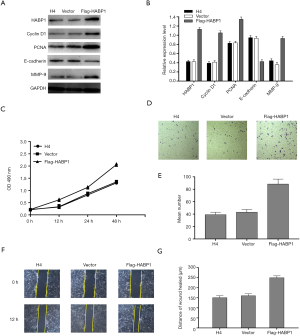
Upregulation of HABP1 expression promotes proliferation, invasion and migration of H4 glioma cells
Based on the experimental results in Figure 4, we decided to use the overexpression of HABP1 to further demonstrate our hypothesis. We used upregulated HABP1 to transfect the H4 cell line with low endogenous expression. Western blotting studies showed an increased expression of PCNA, CyclinD1, MMP-9, and a decreased expression of E-cadherin (Figure 5A,B). CCK-8 assay transfected H4 cells, and cell viability was detected at 0, 12, 24, 48 hours; the results indicated that over-expression of HABP1 potently enhanced cell viability (Figure 5C). We used transwell assay to determine the 24-hour cell migration number of the control group, the empty plasmid group, and the over-expression group. The results showed that the cell migration ability of the over-expression group was significantly higher than that of the other two groups (Figure 5D,E). After 24 hours, the wound healing ability of H4 cells with over-expression of HABP1 was significantly higher than that of the control group (Figure 5F,G).
Discussion
The extracellular matrix (ECM) protects the tissue structure to regulate gene expression under normal conditions, and creates a favorable microenvironment for cancer cells under pathophysiological conditions, regulating tumor formation and development (20). HABP1 may be one of the pathogenesis factors of many ECM tumors (21). It has been reported that HABP1 is involved in different functional processes of cells, including the immune response, splicing mechanism, cell differentiation, and epithelial mesenchymal transition. Due to the cDNA sequence of HABP1 being in line with the cDNA sequence of the p32 protein (22), it has been reported that the expression of HABP1 is associated with poor staging and prognosis in patients with breast cancer and gastric cancer. Furthermore, HABP1 is ubiquitous in glial cells and epithelial cells, suggesting that regulation of the HABP1/p32/gC1qR-mediated pathway may be a marker for the diagnosis of cancer.
We speculated that HABP1 might also regulate the progression of malignant glioma. Therefore, we used fresh glioma tissues to analyze the expression of HABP1 in different grades of glioma tissues by Western blot analysis. It was found that the glioma grade was increased, HABP1 expression was enhanced. Then, we used immunohistochemical staining to evaluate the expression of HABP1 tumor proliferation index, and it was found that Ki-67 expression was positively correlated with HABP1. Therefore, we speculated that HABP1 might be involved in the cytogenesis of glioma.
To demonstrate this hypothesis, we selected five glioma cell lines and found that HABP1 was highly expressed in the U87MG and U251MG cell lines while being relatively low in the H4 cell line. We then screened the interference sites with obvious interference efficiency by transfection and analyzed the expression of PCNA, CyclinD1, E-cadherin, and MMP-9 after HABP1 interference. We found that HABP1 had significant proliferation and invasion effects on glioma. In order to confirm this assumption, we used the overexpression of HABP1, which is the equivalent to the interference with HABP1. The conclusions and speculations were consistent. Undoubtedly, this again proves the migration, proliferation, and invasive effects of HABP1 on malignant glioma.
Conclusions
Generally speaking, this study showed that HABP1 is expressed in human glioma tissues and is positively correlated with the WHO tumor grade. The development of glioma is affected by knocking down or overexpressing HABP1. These data suggest that HABP1 may play an important role in glioma progression. Many studies have shown that HABP1/p32/gC1qR is involved in the regulation of a variety of cellular functions, such as SF2/SRSF1 as a regulator of the splicing mechanism of HABP1 (23), transforming growth factor B (TGFβ) (24), enhancing epithelial cells by up-regulating hyaluronan synthase 2 (Has2), and assisting in cancer cell metastasis through mesenchymal transfer (EMT) (25,26). However, the mechanism for controlling each specific pathway of HABP1 has not been well studied. HABP1 binding affinity ligands, which may be used in glioma therapy, need to be further studied.
Table S1
| Variables | Total | HABP1 expression | χ2 | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age | 0.978 | 0.082 | |||
| <45 | 27 | 10 | 17 | ||
| ≥45 | 53 | 32 | 21 | ||
| Gender | 3.028 | 0.323 | |||
| Men | 33 | 20 | 13 | ||
| Women | 47 | 22 | 25 | ||
| Surgery | 1.626 | 0.202 | |||
| Partial resection | 49 | 29 | 20 | ||
| Gross total resection | 31 | 13 | 18 | ||
| Tumor diameter | 1.776 | 0.183 | |||
| <4 cm | 41 | 25 | 16 | ||
| ≥4 cm | 39 | 17 | 22 | ||
| Tumor location | 1.982 | 0.576 | |||
| Frontal | 22 | 9 | 13 | ||
| Parietal | 22 | 12 | 10 | ||
| Occipital | 15 | 8 | 7 | ||
| Temporal | 21 | 13 | 8 | ||
| WHO grade | 6.790 | 0.034* | |||
| II | 26 | 18 | 8 | ||
| III | 23 | 13 | 10 | ||
| IV | 31 | 11 | 20 | ||
Statistical analyses were performed by the Pearson χ2 test. *, P values <0.05 was considered significant. WHO, World Health Organization.
Table S2
| Characteristic | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| Age | 1.103 | 0.675–2.244 | 0.513 |
| Gender | 1.522 | 0.712–2.279 | 0.115 |
| Surgery | 0.986 | 0.334–2.561 | 0.923 |
| Tumor diameter | 0.597 | 0.276–1.137 | 0.109 |
| Tumor location | 1.306 | 0.598–2.610 | 0.521 |
| WHO grade | 1.473 | 0.771–2.234 | 0.012* |
| HABP1 | 1.387 | 0.665–2.639 | 0.177 |
*, P values <0.05 was considered significant. CI, confidence interval; WHO, World Health Organization.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This research was approved by the Ethics Committee of Nantong University (approval ID: 20190313). Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Peckham-Gregory EC, Montenegro RE, Stevenson DA, et al. Evaluation of racial disparities in pediatric optic pathway glioma incidence: Results from the Surveillance, Epidemiology, and End Results Program, 2000–2014. Cancer Epidemiol 2018;54:90-4. [Crossref] [PubMed]
- Applebaum AJ, Buda K, Kryza-Lacombe M, et al. Prognostic awareness and communication preferences among caregivers of patients with malignant glioma. Psychooncology 2018;27:817-23. [Crossref] [PubMed]
- Krauze AV, Attia A, Braunstein S, et al. Correction to expert consensus on re-irradiation for recurrent glioma. Radiat Oncol 2018;13:8. [Crossref] [PubMed]
- Dogan I, Ucer M, Başkaya MK. Gross Total Resection of Chordoid Glioma of the Third Ventricle via Anterior Interhemispheric Transcallosal Transforaminal Approach at Two Stages. J Neurol Surg B Skull Base 2018;79:S281-S2. [Crossref] [PubMed]
- Metellus P, Coulibaly B, Colin C, et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathologica 2010;120:719-29. [Crossref] [PubMed]
- Ghotme KA, Barreto GE, Echeverria V, et al. Gliomas: New Perspectives in Diagnosis, Treatment and Prognosis. Curr Top Med Chem 2017;17:1438-47. [Crossref] [PubMed]
- Cho DH, Kim YS, Jo DS, et al. Pexophagy: Molecular Mechanisms and Implications for Health and Diseases. Mol Cells 2018;41:55-64. [PubMed]
- Bharadwaj A, Ghosh I, Sengupta A, et al. Stage-specific expression of proprotein form of hyaluronan binding protein 1 (HABP1) during spermatogenesis in rat. Mol Reprod Dev 2002;62:223-32. [Crossref] [PubMed]
- Biswas AK, Hafiz A, Banerjee B, et al. Plasmodium falciparum uses gC1qR/HABP1/p32 as a receptor to bind to vascular endothelium and for platelet-mediated clumping. PLoS Pathog 2007;3:1271-80. [Crossref] [PubMed]
- Prakash M, Kale S, Ghosh I, et al. Hyaluronan-binding protein 1 (HABP1/p32/gC1qR) induces melanoma cell migration and tumor growth by NF-kappa B dependent MMP-2 activation through integrin α(v)β(3) interaction. Cell Signal 2011;23:1563-77. [Crossref] [PubMed]
- Kaul R, Saha P, Saradhi M, et al. Overexpression of hyaluronan-binding protein 1 (HABP1/p32/gC1qR) in HepG2 cells leads to increased hyaluronan synthesis and cell proliferation by up-regulation of cyclin D1 in AKT-dependent pathway. J Biol Chem 2012;287:19750-64. [Crossref] [PubMed]
- Kumar R, Choudhury NR, Salunke DM, et al. Evidence for clustered mannose as a new ligand for hyaluronan-binding protein (HABP1) from human fibroblasts. J Biosci 2001;26:325-32. [Crossref] [PubMed]
- Basu K, Sen A, Ray K, et al. Genetic association and gene-gene interaction of HAS2, HABP1 and HYAL3 implicate hyaluronan metabolic genes in glaucomatous neurodegeneration. Dis Markers 2012;33:145-54. [Crossref] [PubMed]
- Steffens B, Feckler C, Palme K, et al. The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J 2002;27:591-9. [Crossref] [PubMed]
- Ghosh I, Chowdhury AR, Rajeswari MR, et al. Differential expression of Hyaluronic Acid Binding protein 1 (HABP1)/P32/C1QBP during progression of epidermal carcinoma. Mol Cell Biochem 2004;267:133-9. [Crossref] [PubMed]
- Gómezmacías GS, Garzarodríguez ML, Garzaguajardo R, et al. Overexpression of the matrix metalloproteinase 11 gene is a potential biomarker for type 1 endometrial cancer. Oncol Lett 2018;16:1073-8. [PubMed]
- Deng X, Zheng H, Li D, et al. MicroRNA-34a regulates proliferation and apoptosis of gastric cancer cells by targeting silent information regulator 1. Exp Ther Med 2018;15:3705-14. [PubMed]
- Wang F, Li B, Fu P, et al. Immunomodulatory and enhanced antitumor activity of a modified thymosin α1 in melanoma and lung cancer. Int J Pharm 2018;547:611-20. [Crossref] [PubMed]
- Cui H, Zheng M, Zhao G, et al. Identification of differentially expressed genes and pathways for intramuscular fat metabolism between breast and thigh tissues of chickens. BMC Genomics 2018;19:55. [Crossref] [PubMed]
- de Mello LEB, Araujo AN, Alves CX, et al. The G534E variant in HABP2 is not associated with increased risk of familial nonmedullary thyroid cancer in Brazilian Kindreds. Clin Endocrinol (Oxf) 2017;87:113-4. [Crossref] [PubMed]
- Colombo C, Muzza M, Proverbio MC, et al. Segregation and expression analyses of hyaluronan-binding protein 2 (HABP2): insights from a large series of familial non-medullary thyroid cancers and literature review. Clin Endocrinol (Oxf) 2017;86:837. [Crossref] [PubMed]
- Biswas AK, Hafiz A, Banerjee B, et al. Plasmodium falciparumUses gC1qR/HABP1/p32 as a Receptor to Bind to Vascular Endothelium and for Platelet-Mediated Clumping. PLoS Pathog 2007;3:1271-80. [Crossref] [PubMed]
- Kim BC, Hwang HJ, An HT, et al. Antibody neutralization of cell-surface gC1qR/HABP1/SF2-p32 prevents lamellipodia formation and tumorigenesis. Oncotarget 2016;7:49972-85. [PubMed]
- Evanko SP, Potter-Perigo S, Petty LJ, et al. Hyaluronan Controls the Deposition of Fibronectin and Collagen and Modulates TGF-β1 Induction of Lung Myofibroblasts. Matrix Biol 2015;42:74-92. [Crossref] [PubMed]
- Basu K, Sen A, Ray K, et al. Genetic Association and Gene-gene interaction of HAS2, HABP1 and HYAL3 Implicate Hyaluronan Metabolic Genes in Glaucomatous Neurodegeneration. Dis Markers 2012;33:145-54. [Crossref] [PubMed]
- Preca BT, Bajdak K, Mock K, et al. A novel ZEB1/HAS2 positive feedback loop promotes EMT in breast cancer. Oncotarget 2017;8:11530-43. [Crossref] [PubMed]


