
Cyclin-dependent kinase 6 (CDK6) is a candidate diagnostic biomarker for early non-small cell lung cancer
Introduction
Primary lung cancer is the most common cancer, and is the leading cause of cancer-related deaths worldwide, especially in China (1). As we know, non-small cell lung cancer (NSCLC) is the main histological type, accounting for approximately 85% of all lung cancers (2). According to pathological patterns, NSCLC can be mainly divided into lung squamous cell carcinoma (SCC) and adenocarcinoma (AD), accounting for 60–85% of all NSCLC cases. Numerous differences have been reported in the biological behavior and characteristics of lung SCC and AD. For example, AD is more likely to occur in women and non-smokers; whereas SCC is more prevalent in elderly men and is strongly associated with smoking (3). Additionally, several studies based on the Japanese population showed that patients with lung SCC had a worse prognosis than those with AD (4,5). Another Japanese lung cancer registry study based on a larger sample reported a contrary result, showing that AD had a better clinical outcome in comparison with SCC (6).
The cell cycle, a process through which cells duplicate themselves, is frequently disordered during tumor development (7). Generally, cell cycle progression is a highly ordered and tightly-regulated process involving multiple checkpoints that assess extracellular growth signals, cell size, and DNA integrity (8). It is divided into four main phases: G1 phase of cell growth, S phase of DNA replication, followed by G2 phase synthesizing additional proteins for the following cell division phase called M phase. The cell cycle is predominantly regulated by cyclin-dependent kinases (CDKs), cyclins and cyclin-dependent kinase inhibitors (CKIs). CDKs are the catalytic subunits of heterodimeric serine/threonine protein kinases whose best-characterized members are involved in controlling progression through the cell cycle (9,10).
CDK6, a member of CDK family, can specifically bind to cyclin D and phosphorylate the retinoblastoma 1 protein, thereby regulating the progression of cell cycle from G1 to S phase (11). A previous study showed that CDK6 could be activated in the initial phase of cell cycle progression by specific binding to cyclin D to facilitate cell transition in the G1 phase (12). On the other hand, CDK6 could inhibit the catalytic activity of the cyclin D-CDK6 complex by its specific binding to p16, thereby blocking the cell cycle at the G1 phase (13). Abnormal expression of CDK6 has been reported in a variety of cancers, such as bladder cancer, lymphoma, leukemia and a small subset of malignant gliomas (14-18). Moreover, Kollmann et al. reported that CDK6 could exert its full tumor-promoting function by enhancing proliferation and stimulating angiogenesis in lymphoid malignancies (19). It has been reported that CDK6 participated in cell proliferation and apoptosis of NSCLC cells (20,21). Furthermore, the inhibitors of CDK6 were promising agents in the treatment of NSCLC (22). However, to date, limited research has been reported about the relationship between CDK6 and NSCLC, especially in the T1 stage. In this study, we examined the CDK6 expression in cases of NSCLC and the distal normal tissues using real-time PCR (RT-PCR) and immunohistochemical staining, and determined the relationship between CDK6 expression and the clinicopathological features of T1 stage NSCLC.
Methods
Patients and tissues
Primary lung cancer specimens with T1 stage were collected from 56 Chinese patients that underwent surgery and had detailed clinicopathological data in the biobank of Zhejiang Cancer Hospital from August 2008 to October 2012. Those with multiple lesions, other carcinoma or had been pretreated with radiotherapy or chemotherapy were excluded. There were no sex and age restrictions. The American Joint Committee on Cancer (AJCC) and World Health Organization (WHO) guidelines were used to determine the tumor staging. Each patient signed a written informed consent to participate in the study prior to surgery. This study was approved by the Medical Ethics Committee, Zhejiang Cancer Hospital.
Two pairs of tumor tissue and distal normal tissue (>5 cm from tumor) were obtained from each patient. The tissues were quickly frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Other pieces of the tissue were embedded into paraffin blocks for paraffin sectioning.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen sections of tumor and distal normal samples using NucleoSpin® TriPrep kit (MACHEREY-NAGEL, Germany) according to the protocol. RNA purity and concentration were determined using a NanoDrop 1000 (Thermo Scientific, USA). Total RNA (1 µg) was used for cDNA synthesis using the PrimeScriptTM Reagent Kit (TaKaRa, China) according to the manufacturer’s instructions.
RT-PCR analysis
Amplification and melt curve analysis were performed using an ABI 7500 PCR system (Applied Biosystems, USA). Reactions were carried out in a total volume of 20 µL using SYBR® Premix Ex Taq Kit (TaKaRa). PCR primers used were as follows: CDK6, 5'-TTCCCAGGCAGGCTTTTCAT-3' (forward) and 5'-CTGTATTCAGCTCCGAGGTGTTCT-3' (reverse); β-actin, 5'-TGGCACCCAGCACAATGAA-3' (forward) and 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3' (reverse).
PCR reactions were performed as follows: initial denaturation at 95 °C for 1 min, 40 amplification cycles of denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 1 min. Each PCR reaction used one premixed template. For expression analysis, β-actin was used as the internal control, and the relative quantification of gene in test tissues was calculated using the following equation: amount of target gene =2-ΔΔCt; ΔΔCt = [Ct (gene, test) − Ct (ACTB, test)] − [Ct (gene, reference) − Ct (ACTB, reference)] (23). To confirm the specificity and accuracy of the PCR reaction, PCR products were electrophoresed on a 2% agarose gel and sequenced (Shanghai GeneCore BioTechnologies Co, Ltd., Shanghai, China). All PCR reactions were performed in triplicate.
Immunohistochemical staining
The immunohistochemical staining kit was purchased from Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd. Paraffin-embedded tissues were cut to 4 µm thickness and placed at 72 °C for 30 min, then dewaxed and hydrated using xylene and ethanol. Antigens were heat retrieved with citrate buffer. After three washes, the slides were placed in a 3% hydrogen peroxide solution for 5–10 min and then washed with running water twice and PBS for 5 min. The primary antibodies (rabbit monoclonal antibody, Abcam, Cambridge, UK) were added (1:100 dilution) and incubated overnight at 4 °C. After washing with water twice and PBS for 5 min, the secondary antibodies were added and incubated for 20 min, followed by washing, DAB staining, counterstaining, and mounting.
Evaluation of immunohistochemical staining
The immunohistochemically stained tissue sections were judged independently by two pathologists. The expression of CDK6 detected by immunohistochemistry was evaluated by measuring staining intensity and determining the proportion of positively-stained cells. The nuclear and cytoplasm were scored separately, using the scoring method described previously (24). For cytoplasmic staining, the score was evaluated according to the staining intensity, there were four levels including negative (0 point), weak positive (1 point), intermediate positive (2 points), and strong positive (3 points). The proportion of positive cells was scored as follows: <5% was 0 point, 5 30% was 1 point, 31–75% was 2 points, and >75% was 3 points. The sum of the two fractions was the cytoplasmic score. The nuclear staining intensity score was consistent with the cytoplasm and positive nuclear staining scores were defined as follows: <10% was 0 point, 10–50% was 1 point, 51–80% was 2 points, and >80% was 3 points. The sum of the cytoplasm and nuclear staining scores was used as the final staining score for CDK6 (0–12). If the sum of two scores was <6 points, it was considered negative; and ≥6 points indicated positive expression.
Statistical analysis
The expression of CDK6 in tumor tissues and normal tissues was compared using the paired samples T-test and nonparametric tests. The relationship between tumor tissues (or normal tissues) and clinical pathological factors was analyzed with the independent samples t-test. The criterion for statistical significance was set at P<0.05 and statistical analysis was performed with SPSS 13.0 software (IBM, Chicago, IL, USA).
Results
Clinicopathological features of T1 stage NSCLC
A total of 56 pairs of tumor and distal normal tissue samples were collected from patients with T1 stage NSCLC for target gene examination and immunohistochemical staining. Of the 56 patients, 40 (71.4%) were male and 16 (28.6%) were female. The median age was 62 years (range, 38–79 years), and 55.4% of the patients had a smoking history of greater than or equal to 20 pack-years. In terms of the pathological type, more than half of the patients (55.4%) had SCC and 44.6% had AD. Twenty-seven patients (48.2%) were in T1a stage (tumor diameter ≤2 cm) and 29 patients (51.8%) were in T1b stage (tumor diameter 2–3 cm). Lymph node metastasis was found in 19.6% of the patients.
The expression of CDK6 in T1 stage NSCLC
RT-PCR and immunohistochemical staining were used to explore the expression levels of CDK6 in the 56 cases of T1 stage NSCLC. The relative quantification of CDK6 in distal normal tissues and tumor tissues was 0.648±0.301 and 0.853±0.776, respectively (P=0.073). There was a trend of increased expression of CDK6 mRNA in tumor tissues (Table 1). The expression of CDK6 protein in the 56 pairs of NSCLC tissue samples and normal lung tissue samples was also detected by immunohistochemical staining (Figure 1). CDK6 protein expression was found to be positive in 8 NSCLC samples (14.3%), whereas it was negative in all distal normal tissue samples (100%). Moreover, nonparametric tests showed that the expression of CDK6 protein in NSCLC tissues was significantly higher than that in normal lung tissues (P=0.003).
Table 1
| Tissues | N | M | SD | t | P* |
|---|---|---|---|---|---|
| Normal | 56 | 0.648 | 0.301 | – | – |
| Tumor | 56 | 0.853 | 0.776 | – | – |
| Tumor minus normal | 56 | 0.205 | 0.838 | 1.830 | 0.073 |
*, independent samples t-test. CDK6, Cyclin-dependent kinase 6; NSCLC, non-small cell lung cancer.

Correlation between CDK6 expression and clinicopathological features in NSCLC
We then examined the correlation between CDK6 gene expression and clinicopathological features of T1 stage NSCLC patients including sex (male versus female), age (<60 versus ≥60 years), smoke index (<20 versus ≥20 pack-years), tumor diameter (≤2 versus 2–3 cm), pathological type (SCC versus AD), differentiation (poorly versus well-moderately), visceral pleura involvement (no versus yes), vascular tumor thrombus (no versus yes) and lymph node metastasis (no versus yes) (Table 2). CDK6 gene expression was associated with clinicopathological features including smoking status and pathological types in tumor tissues, while no significant correlation was found between CDK6 gene expression and clinicopathological features in distal normal tissues. With the increase of smoking quantity, CDK6 gene expression was accordingly increased (P=0.009). Furthermore, CDK6 gene expression in SCC was significantly higher than that in AD (P=0.000). Subsequently, we further detected CDK6 protein expression in 31 SCC tissues and 25 AD tissues by immunohistochemical staining (Figure 1), and found that CDK6 protein expression was positive in 8 SCC tissues (25.8%) whereas it was negative in all AD tissues (100%), which indicated that the expression of CDK6 protein was significantly higher in SCC tissues than that in AD tissues (P<0.001). The results of protein level quantification were consistent with those of RT-PCR.
Table 2
| Variables | N | Normal | Tumor | |||||
|---|---|---|---|---|---|---|---|---|
| M | t | P* | M | t | P* | |||
| Gender | ||||||||
| Male | 40 | 0.627±0.311 | −0.863 | 0.395 | 0.962±0.725 | 1.688 | 0.097 | |
| Female | 16 | 0.700±0.274 | 0.581±0.857 | |||||
| Age (years) | ||||||||
| <60 | 22 | 0.560±0.236 | −1.799 | 0.078 | 0.867±0.609 | 0.110 | 0.913 | |
| ≥60 | 34 | 0.705±0.327 | 0.844±0.877 | |||||
| Smoke index (pack-years) | ||||||||
| <20 | 25 | 0.713±0.300 | 1.475 | 0.146 | 0.557±0.726 | −2.706 | 0.009 | |
| ≥20 | 31 | 0.595±0.295 | 1.092±0.742 | |||||
| Tumor diameter (cm) | ||||||||
| ≤2 | 27 | 0.626±0.291 | −0.528 | 0.600 | 0.719±0.778 | −1.250 | 0.217 | |
| 2–3 | 29 | 0.669±0.313 | 0.977±0.767 | |||||
| Pathological type | ||||||||
| SCC | 31 | 0.595±0.273 | −1.487 | 0.143 | 1.265±0.815 | 5.992 | 0.000 | |
| AD | 25 | 0.714±0.325 | 0.342±0.243 | |||||
| Differentiation | ||||||||
| Poorly | 21 | 0.655±0.326 | 0.128 | 0.899 | 0.742±0.467 | −0.956 | 0.343 | |
| Well-moderately | 35 | 0.644±0.289 | 0.919±0.913 | |||||
| Visceral pleura involvement | ||||||||
| No | 48 | 0.657±0.310 | 0.519 | 0.606 | 0.909±0.815 | 1.337 | 0.187 | |
| Yes | 8 | 0.597±0.250 | 0.516±0.351 | |||||
| Vascular tumor thrombus | ||||||||
| No | 52 | 0.656±0.296 | 0.724 | 0.472 | 0.857±0.793 | 0.152 | 0.880 | |
| Yes | 4 | 0.543±0.387 | 0.796±0.598 | |||||
| Lymph node metastasis | ||||||||
| No | 45 | 0.651±0.315 | 0.157 | 0.876 | 0.912±0.818 | 1.163 | 0.250 | |
| Yes | 11 | 0.635±0.247 | 0.610±0.537 | |||||
*, independent samples t-test. CDK6, Cyclin-dependent kinase 6; SCC, squamous cell carcinomas; AD, adenocarcinoma.
The differential expression of CDK6 in different pathological types of NSCLC
Cases were stratified by tumor pathological types to explore whether CDK6 gene expression was associated with NSCLC in this selected population of patients. As shown in Table 3, CDK6 gene expression was increased in SCC tissues (1.265±0.815) compared with that in the distal normal tissues (0.595±0.273) and the difference was significant (P=0.000). Nonparametric tests also showed that the expression of CDK6 protein in SCC tissues was significantly higher than that in distal normal tissues (P<0.001). However, reduced expression of CDK6 was detected in AD and the relative quantification of CDK6 gene in distal normal tissues and tumor tissues was 0.714±0.325 and 0.342±0.243, respectively (P=0.000).
Table 3
| Pathological type | Tissues | N | M | SD | t | P* |
|---|---|---|---|---|---|---|
| SCC | Normal | 31 | 0.595 | 0.273 | – | – |
| Tumor | 31 | 1.265 | 0.815 | – | – | |
| Tumor minus normal | 31 | 0.67 | 0.808 | 4.617 | 0.000 | |
| AD | Normal | 25 | 0.714 | 0.325 | – | – |
| Tumor | 25 | 0.342 | 0.243 | – | – | |
| Tumor minus normal | 25 | −0.372 | 0.407 | −4.572 | 0.000 |
*, Paired samples t-test. CDK6, Cyclin-dependent kinase 6; SCC, squamous cell carcinomas; AD, adenocarcinoma.
Correlation between CDK6 expression and clinicopathological features in different pathological types of NSCLC
The correlation between CDK6 gene expression and clinicopathological features in SCC and AD were then analyzed. No clinicopathological features were found to be related to CDK6 gene expression in SCC (P>0.05) (Table 4). However, the expression of the CDK6 gene in AD was negatively correlated with tumor diameter (P=0.049) (Table 5). Compared with distal normal tissues, CDK6 gene expression was decreased significantly in tumor tissues no matter whether the tumor diameter was less than or larger than 2 cm in patients with lung AD (P<0.05; Table S1).
Table 4
| Variables | N | M | SD | t | P* |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 28 | 1.227 | 0.703 | −0.387 | 0.735 |
| Female | 3 | 1.621 | 1.751 | ||
| Age (years) | |||||
| <60 | 14 | 1.173 | 0.555 | −0.565 | 0.577 |
| ≥60 | 17 | 1.341 | 0.990 | ||
| Smoke index (pack-years) | |||||
| <20 | 5 | 1.416 | 1.295 | 0.445 | 0.660 |
| ≥20 | 26 | 1.237 | 0.723 | ||
| Tumor diameter (cm) | |||||
| ≤2 | 9 | 1.394 | 1.019 | 0.556 | 0.583 |
| 2–3 | 22 | 1.213 | 0.736 | ||
| Differentiation | |||||
| Poorly | 12 | 1.051 | 0.376 | 1.398 | 0.174 |
| Well-moderately | 19 | 1.401 | 0.985 | ||
| Vascular tumor thrombus | |||||
| No | 28 | 1.297 | 0.837 | 0.655 | 0.517 |
| Yes | 3 | 0.970 | 0.595 | ||
| Lymph node metastasis | |||||
| No | 29 | 1.251 | 0.838 | −0.364 | 0.719 |
| Yes | 2 | 1.471 | 0.399 |
*, independent samples t-test. CDK6, Cyclin-dependent kinase 6; SCC, squamous cell carcinomas.
Table 5
| Variables | N | M | SD | t | P* |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 12 | 0.343 | 0.221 | 0.021 | 0.983 |
| Female | 13 | 0.341 | 0.271 | ||
| Age (years) | |||||
| <60 | 8 | 0.332 | 0.153 | −0.136 | 0.893 |
| ≥60 | 17 | 0.346 | 0.280 | ||
| Smoke index (pack-years) | |||||
| <20 | 20 | 0.342 | 0.264 | 0.028 | 0.978 |
| ≥20 | 5 | 0.339 | 0.150 | ||
| Tumor diameter (cm) | |||||
| ≤2 | 18 | 0.382 | 0.275 | 2.086 | 0.049 |
| 2–3 | 7 | 0.238 | 0.063 | ||
| Differentiation | |||||
| Poorly | 9 | 0.331 | 0.136 | 0.187 | 0.854 |
| Well-moderately | 16 | 0.347 | 0.291 | ||
| Visceral pleura involvement | |||||
| No | 17 | 0.260 | 0.111 | −2.013 | 0.080 |
| Yes | 8 | 0.516 | 0.351 | ||
| Lymph node metastasis | |||||
| No | 16 | 0.298 | 0.169 | −1.002 | 0.339 |
| Yes | 9 | 0.418 | 0.337 |
*, independent samples t-test. CDK6, Cyclin-dependent kinase 6; AD, adenocarcinoma.
We also found a related trend between expression of the CDK6 gene and visceral pleura involvement in AD (P=0.080). When stratified further, CDK6 gene expression in tumor tissues was significantly lower than that in distal normal tissues for AD without visceral pleura involvement (P=0.000). In contrast, there was no statistically significant difference in CDK6 gene expression between tumor and normal tissues in cases with visceral pleura involvement (P=0.559; Table S2).
Discussion
As we know, it is a common malignant behavior of human cancer that cell cycle deregulation results in uncontrolled cell proliferation. Cell cycle progression is regulated by a subfamily of CDKs, which belongs to the core cell cycle machinery and comprises a family of serine/threonine kinase (14,25,26). CDK6, an important cell cycle regulatory factor, plays a significant role in cell proliferation, differentiation and angiogenesis while driving the G1 phase progression and the transition of the cell cycle from G1 to S phase (19,27-29). CDK6 has been proven to bind to and enhance degradation of the eyes absent homolog 2 protein (EYA2), which is necessary for the development of multiple organs, regulation of cell proliferation, and is disordered in various cancers. This interaction suggests that the mechanism by which CDK6 regulates EYA2 activity could be important in development and in cancer (27).
Kollmann et al. has reported that the proliferation and cell cycle progression can be inhibited by CDK6, which is overexpressed in BCR-ABL transformed B cell leukemia/lymphoma cells (19). CDK6 overexpression also has been shown to block differentiation of leukemic cells and osteoblasts (15,30,31). Matushansky et al. found that there was a negative regulatory relationship between the expression of CDK6 and the differentiation of murine erythroid leukemia (MEL) cells. When MEL cells were induced to re-enter differentiation, CDK6 was downregulated (28). Kollmann et al. have shown that CDK6 is overexpressed in lymphoma and regulates the transcription of vascular endothelial growth factor A (VEGFA) that promotes tumor angiogenesis and controls the formation of neovascularization, which indicates that CDK6 not only plays a role in the cell cycle, but also regulates angiogenesis in lymphoma, and further confirms the critical role of CDK6 in hematopoietic malignancies (19). Additionally, amplification and overexpression of CDK6 has also been detected in a variety of solid tumors, e.g., neuroblastomas, oral SCCs, breast tumors, sarcomas and melanomas (27,32-35).
In this study, we showed that CDK6 had a tendency to increase in tumor tissues compared to the transcriptional level in normal tissues, although it did not reach significance. However, the expression of CDK6 protein in NSCLC tissues was significantly higher than that in normal lung tissues, which was consistent with the results as reported by Hong et al. (36). Igarashi et al. reported that CDK6 proteins were significantly increased in lung cancer tissues in comparison to adjacent non-cancerous tissues. Stratified by pathological types, CDK6 protein was significantly increased in lung ADs, but no significant change was found in lung SCCs (37). In contrast, we found that CDK6 was overexpressed in SCC, however, in AD CDK6 showed significantly lower expression than in normal tissues. The TCGA database also confirmed that the CDK6 expression in AD was lower than that in normal tissue and was higher than that in normal tissue in SCC (Figure S1). The difference may be because our study population focused on those with T1 stage NSCLC, while in the small sample study of Igarashi et al., most patients with NSCLC were in stage II and III, without subgroup analysis of early lung cancer alone. Particularly, we found CDK6 gene expression was not associated with any clinicopathological features in SCC, whereas it was negatively correlated with tumor diameter in AD. CDK6 as an oncogene, promotes cell proliferation in many kinds of cancers, while overexpression of CDK6 has also been shown to reduce skin tumorigenesis (38-41). We suspected that this dual effect of CDK6 led to this result. Of course, our research only described this phenomenon. The reasons and mechanisms driving this need to be further investigated. In addition, we found that the expression of the CDK6 gene had a correlation with visceral pleura involvement in AD, suggesting that CDK6 may have the potential to be a diagnostic indicator for AD, but not for SCC.
The expression of CDK6 was increased in pulmonary SCC but decreased in pulmonary AD with T1 stage, and the clinicopathological implications of CDK6 expression in AD and SCC were found to be different, which provides strong support for to study CDK6 to better understand and further scrutinize the complexity of lung cancer.
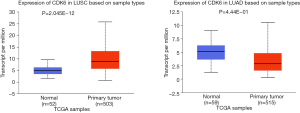
Table S1
| Tumor diameter | Tissues | N | M | SD | t | P* |
|---|---|---|---|---|---|---|
| ≤2 cm | Normal | 18 | 0.685 | 0.304 | ||
| Tumor | 18 | 0.382 | 0.275 | |||
| Tumor minus normal | 18 | −0.303 | 0.401 | −3.206 | 0.005 | |
| 2–3 cm | Normal | 7 | 0.788 | 0.390 | ||
| Tumor | 7 | 0.238 | 0.063 | |||
| Tumor minus normal | 7 | −0.551 | 0.395 | −3.686 | 0.010 |
*, Paired samples
Table S2
| Visceral pleura involvement | Tissues | N | M | SD | t | P* |
|---|---|---|---|---|---|---|
| No | Normal | 17 | 0.769 | 0.349 | ||
| Tumor | 17 | 0.26 | 0.111 | |||
| Tumor minus normal | 17 | −0.509 | 0.354 | −5.937 | 0.000 | |
| Yes | Normal | 8 | 0.597 | 0.25 | ||
| Tumor | 8 | 0.516 | 0.351 | |||
| Tumor minus normal | 8 | −0.081 | 0.374 | −0.613 | 0.559 |
*, Paired samples
Acknowledgments
Funding: Our study was supported financially by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by Medical Ethics Committee, Zhejiang Cancer Hospital. The American Joint Committee on Cancer (AJCC) and World Health Organization (WHO) guidelines were used to determine the tumor staging. Each patient signed a written informed consent to participate in the study prior to surgery.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung cancers: Molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 2018; [Crossref] [PubMed]
- Fukui T, Taniguchi T, Kawaguchi K, et al. Comparisons of the clinicopathological features and survival outcomes between lung cancer patients with adenocarcinoma and squamous cell carcinoma. Gen Thorac Cardiovasc Surg 2015;63:507-13. [Crossref] [PubMed]
- Ichinose Y, Yano T, Asoh H, et al. Prognostic factors obtained by a pathologic examination in completely resected non-small-cell lung cancer. An analysis in each pathologic stage. J Thorac Cardiovasc Surg 1995;110:601-5. [Crossref] [PubMed]
- Suzuki K, Nagai K, Yoshida J, et al. Conventional clinicopathologic prognostic factors in surgically resected nonsmall cell lung carcinoma. A comparison of prognostic factors for each pathologic TNM stage based on multivariate analyses. Cancer 1999;86:1976-84. [Crossref] [PubMed]
- Asamura H, Goya T, Koshiishi Y, et al. A Japanese Lung Cancer Registry study: prognosis of 13,010 resected lung cancers. J Thorac Oncol 2008;3:46-52. [Crossref] [PubMed]
- Dominguez-Brauer C, Thu KL, Mason JM, et al. Targeting Mitosis in cancer: Emerging strategies. Mol Cell 2015;60:524-36. [Crossref] [PubMed]
- Park MT, Lee SJ. Cell cycle and cancer. J Biochem Mol Biol 2003;36:60-65. [PubMed]
- Swaffer MP, Jones AW, Flynn HR, et al. CDK substrate phosphorylation and ordering the cell cycle. Cell 2016;167:1750-1761.e16. [Crossref] [PubMed]
- Kumar A, Gopalswamy M, Wolf A, et al. Phosphorylation-induced unfolding regulates p19(INK4d) during the human cell cycle. Proc Natl Acad Sci U S A 2018;115:3344-9. [Crossref] [PubMed]
- Mi D, Manuel M, Huang YT, et al. Pax6 lengthens G1 phase and decreases oscillating Cdk6 levels in murine embryonic cortical progenitors. Front Cell Neurosci 2018;12:419. [Crossref] [PubMed]
- Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol 1994;14:2077-86. [Crossref] [PubMed]
- Russo AA, Tong L, Lee JO, et al. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 1998;395:237-43. [Crossref] [PubMed]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer 2009;9:153-66. [Crossref] [PubMed]
- Chilosi M, Doglioni C, Yan Z, et al. Differential expression of cyclin-dependent kinase 6 in cortical thymocytes and T-cell lymphoblastic lymphoma/leukemia. Am J Pathol 1998;152:209-17. [PubMed]
- Lien HC, Lin CW, Huang PH, et al. Expression of cyclin-dependent kinase 6 (cdk6) and frequent loss of CD44 in nasal-nasopharyngeal NK/T-cell lymphomas: comparison with CD56-negative peripheral T-cell lymphomas. Lab Invest 2000;80:893-900. [Crossref] [PubMed]
- Costello JF, Plass C, Arap W, et al. Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res 1997;57:1250-4. [PubMed]
- Wang G, Zheng L, Yu Z, et al. Increased cyclin-dependent kinase 6 expression in bladder cancer. Oncol Lett 2012;4:43-6. [Crossref] [PubMed]
- Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 2013;24:167-81. [Crossref] [PubMed]
- Fan YF, Yu ZP, Cui XY. lncRNA colorectal neoplasia differentially Expressed (CRNDE) promotes proliferation and inhibits apoptosis in non-small cell lung cancer cells by regulating the miR-641/CDK6 axis. Med Sci Monit 2019;25:2745-55. [Crossref] [PubMed]
- Liu Z, Lu C, Zhao G, et al. Downregulation of miR-218 by nicotine promotes cell proliferation through targeting CDK6 in non-small cell lung cancer. J Cell Biochem 2019;120:18370-7. [PubMed]
- Liu L, Chen Y, Li Q, et al. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem 2019;120:18736-50. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Wu A, Wu B, Guo J, et al. Elevated expression of CDK4 in lung cancer. J Transl Med 2011;9:38. [Crossref] [PubMed]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999;13:1501-12. [Crossref] [PubMed]
- Tigan AS, Bellutti F, Kollmann K, et al. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene 2016;35:3083-91. [Crossref] [PubMed]
- Kohrt D, Crary J, Zimmer M, et al. CDK6 binds and promotes the degradation of the EYA2 protein. Cell Cycle 2014;13:62-71. [Crossref] [PubMed]
- Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene 2003;22:4143-9. [Crossref] [PubMed]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013;140:3079-93. [Crossref] [PubMed]
- Moro T, Ogasawara T, Chikuda H, et al. Inhibition of Cdk6 expression through p38 MAP kinase is involved in differentiation of mouse prechondrocyte ATDC5. J Cell Physiol 2005;204:927-33. [Crossref] [PubMed]
- Scheicher R, Hoelbl-Kovacic A, Bellutti F, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood 2015;125:90-101. [Crossref] [PubMed]
- Andisheh-Tadbir A, Ashraf MJ, Jeiroodi N. Expression of CDK6 in oral squamous cell carcinomas. Asian Pac J Cancer Prev 2018;19:1013-6. [PubMed]
- Dai X, Li L, Liu X, et al. Cooperation of DLC1 and CDK6 affects breast cancer clinical outcome. G3 (Bethesda) 2014;5:81-91. [Crossref] [PubMed]
- Zhu K, Liu L, Zhang J, et al. MiR-29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6. Protein Cell 2016;7:434-44. [Crossref] [PubMed]
- Easton J, Wei T, Lahti JM, Kidd VJ. Disruption of the cyclin D/cyclin-dependent kinase/INK4/retinoblastoma protein regulatory pathway in human neuroblastoma. Cancer Res 1998;58:2624-32. [PubMed]
- Hong JH, Roh KS, Suh SS, et al. The expression of microRNA-34a is inversely correlated with c-MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients. Tumour Biol 2015;36:9327-37. [Crossref] [PubMed]
- Igarashi K, Masaki T, Shiratori Y, et al. Activation of cyclin D1-related kinase in human lung adenocarcinoma. Br J Cancer 1999;81:705-11. [Crossref] [PubMed]
- Tadesse S, Yu M, Kumarasiri M, et al. Targeting CDK6 in cancer: State of the art and new insights. Cell Cycle 2015;14:3220-30. [Crossref] [PubMed]
- Whiteway SL, Harris PS, Venkataraman S, et al. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J Neurooncol 2013;111:113-21. [Crossref] [PubMed]
- Lyu K, Xu Y, Yue H, et al. Long noncoding RNA GAS5 acts as a tumor suppressor in laryngeal squamous cell carcinoma via miR-21. Cancer Manag Res 2019;11:8487-98. [Crossref] [PubMed]
- Gao H, Yin Y, Qian A, et al. LncRNA LINC00974 upregulates CDK6 to promote cell cycle progression in gastric carcinoma. Cancer Biother Radiopharm 2019; [Epub ahead of print]. [Crossref] [PubMed]


