
CENPE promotes glioblastomas proliferation by directly binding to WEE1
Introduction
Use the GLOBOCAN 2018 estimates of cancer incidence and mortality produced by the International Agency for Research on Cancer, there are 296,851 new cancer cases and 241,037 million cancer deaths in brain, nervous system in 2018 (1). Glioblastoma (also known as GBM) is a malignant grade IV tumor in which most tumor cells multiply and divide at any given time. GBM is usually classified as an isocitrate dehydrogenase (IDH)-wild type (WT) or IDH-mutant. IDH-WT GBM accounts for the majority, is more aggressive, and has a worse prognosis. With standard treatment, the median survival of adult GBM (IDH-WT) is approximately 11–15 months (2,3). Therefore, further understanding of the proliferation mechanism of glioma, especially GBM, can play an important role in the control and treatment of GBM.
Cell proliferation is achieved by mitosis. The most important process of mitosis is the cell cycle. The separation of chromosomes is closely related to the correct assembly of microtubules. In addition, microtubules play an important role in maintaining cell shape, cell movement and intracellular organelles movement. Centromere protein E (CENPE) is one of human kinetochore proteins of 312 kDa (4). Studies have shown that knockdown of CENPE or knockdown of xeroderma pigmentosum group A (XPA)-binding protein 2 (XAB2) can inhibit cell proliferation, and CENPE is directly regulated by XAB2 (5). In addition, the expression of CENPE is closely related to the proliferative capacity of tumors. In incurable castration-resistant prostate cancer (CRPC), knockout or pharmacological inhibition of CENPE significantly inhibits tumor growth (6). In gastric cancer cells, when treated with mycophenolic acid (MPA), the expression of CENPE was significantly inhibited, and cell proliferation was inhibited (7). In human neuroblastoma cell lines, knockdown of CENPE results in inhibition of cell proliferation in vitro and in vivo (8).
Methods
Cell lines and patient samples
All of the GBM tumors and paired normal tissue (NT) samples were collected from Tongde Hospital of Zhejiang Province. The study was approved by institutional/regional/national ethics committee/ethics board of Ethics Committee of Zhejiang Institute of Traditional Chinese Medicine (No. XMSC2018048). The U251 and U87 human GBM cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM high glucose medium (Hyclone, USA) with 10% fetal bovine serum (FBS; Hyclone, USA) and 1% penicillin/streptomycin (Gibco, USA), then, incubated at 37 °C in an atmosphere of 5% CO2.
Small interfering RNA (siRNA)
2.5×105 cells were seeded in 6-well plates, cultured overnight (about 12 hours), then, cells were transfected with siRNA (siCENPE, 50 nmol/L) or negative control siRNA (NC, 50 nmol/L) using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) follow the manufacturer’s protocol. The sequence of the siRNA is: CENPE, 5'-CAUCAACUUAUUACGUUAA-3'.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was used to extract total RNA from tissue samples or cells. An RT reagent kit with gDNA Eraser (TaKaRa Biotech, Dalian, China) was used to reverse transcribe RNA into cDNA. The quantitative real-time PCR (qRT-PCR) reaction was then performed using SYBR Green (TaKaRa Biotech, Dalian, China) and detected with ABI 7500 System (Applied Biosystems, Foster City, CA, USA), each sample were repeated three times. All PCR primers were purchased from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA): CENPE: forward, 5'-GATTCTGCCATACAAGGCTACAA-3', reverse, 5'-TGCCCTGGGTATAACTCCCAA-3'; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): forward, 5'-ACAACTTTGGTATCGTGGAAGG-3', reverse, 5'-GCCATCACGCCACAGTTTC-3'. Relative expression levels were calculated using the 2–ΔΔCT method.
Western blot (WB) analysis
Cell and tissue samples were both lysed in RIPA lysis buffer (Beyotime, Haimen, China) containing 1% protease inhibitor PMSF (Beyotime, Haimen, China). Then, a total of 40 micrograms of protein was separated by 10% SDS-PAGE, and the protein was transferred to a PVDF membrane (Roche Applied Sciences, Indianapolis, IN, USA). Membranes were blocked in 5% BSA (Boster, AR0004, Wuhan, China) for 1 hour at room temperature and then incubated overnight at 4 °C with the primary antibody (anti-CENPE, anti-WEE1 anti-GAPDH). After incubation with a suitable HRP-conjugated secondary antibody (Cell Signaling Technology, Danvers, MA, USA) for 1 hour at room temperature, the membrane was visualized with an ECL kit (Beyotime, Haimen, China), and then membranes were visualized using LI-COR Odyssey system (Odyssey; LI-COR Biosciences). The proteins were visualized using a chemiluminescence method (ECL plus WB detection system; Amersham Biosciences, Foster City, CA, USA). The analysis included mouse monoclonal anti-GAPDH (Boster, BM3876, Wuhan, China), rabbit anti-CENPE (Abcam, ab133583, Cambridge, UK), mouse anti-WEE1 (Abcam, ab23354, Cambridge, UK).
Coimmunoprecipitation (co-IP)
After cells were harvested, the 200 µL cell lysate was incubated with ChIP-grade protein G magnetic beads (CST, Danvers, MA, USA) and 5 µL antibodies, shook overnight at 4 °C, wash five times with cold PBS [contain 1% PMSF (Beyotime, Haimen, China)] for 5 minutes each time, suspended in 2× SDS-PAGE loading buffer and then detected by WB with relevant antibodies. The antibodies used for IP were rabbit anti-CENPE (Abcam, ab133583, Cambridge, UK), rabbit anti-WEE1 (Abcam, ab23354, Cambridge, UK).
Cell proliferation assay
The 647-flow cytometry assay kit (GeneCopoeia, Rockville, MD, USA) was used to detect cell proliferation. After the cells were transfected with small RNA for 24 hours, 5-ethynyl-2’-deoxyuridine (EDU) (10 µM) was added to each well for 12 hours, and then stained and flow-detected according to the manufacturer's protocol.
Cell cycle analysis
Propidium iodide (PI) (Beyotime, Haimen, China) staining is used to determine the cell cycle. After 24 hours of cell small RNA interference in the 6-well plate, each well was separately digested and collected into an EP tube. The cells were permeabilized overnight with PBS at 75% cold ethanol, then washed twice with PBS and stained with PI solution for 15 minutes at room temperature [50 µg/mL PI in PBS, 0.1% Triton-X100, 50 µg/mL RNase A (TaKaRa Biotech, Dalian, China)]. Cells were detected within 3 hours using flow cytometer.
Analysis of data from The Cancer Genome Atlas (TCGA)
Difference expression of CENPE in normal and gliomas and survival in glioma patients was analyzed in the website Gliovis (http://gliovis.bioinfo.cnio.es/). And the Graphpad Prism 6 software was used to analyze data.
Results
Expression of CENPE in collected GBM tissues
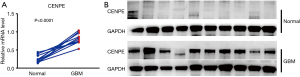
We examined the mRNA expression levels of CENPE in GBM and paracancerous samples by RT-qPCR and analyzed protein expression by WB. mRNA assay results showed that CENPE mRNA levels were significantly elevated in GBM tissues compared to paired normal brain tissue samples (Figure 1A, P<0.0001). WB results showed that the expression level of CENPE in GBM tissues was higher than that in paired NTs, and the expression of CENPE was almost undetectable in paired normal brain tissues (Figure 1B).
Expression of CENPE in TCGA low-grade gliomas and glioblastomas (GBMLGG)
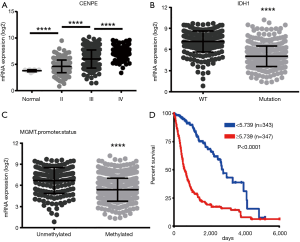
To further determine the expression of CENPE in gliomas, we analyzed data on gliomas in the TCGA database, including GBMLGG. It was found that as the grade of glioma increased, the expression of CENPE also increased (Figure 2A). At the same time, we also found that the expression of CENPE was reduced in GBM with IDH1 mutation (Figure 2B). At the same time, the O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation tissue also showed low CENPE expression (Figure 2C). We then analyzed the survival curves of gliomas with high and low expression of CENPE and found that patients with high expression of CENPE had a worse prognosis (Figure 2D).
CENPE promotes proliferation of glioma cell lines
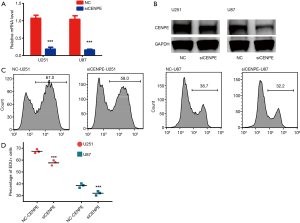
To examine the effect of CENPE on glioma cell proliferation, we performed an EDU assay on CENPE-interfering cell lines. First, we designed siRNA to knock down CENPE in glioma cell lines. CENPE mRNA and protein levels significantly reduced after RNA interference (Figure 3A,B). EDU experiments demonstrated that proliferation of glioma cells was inhibited after CENPE knockdown (Figure 3C,D) (P<0.0001).
Inhibition of CENPE mainly inhibits G0–G1 transition
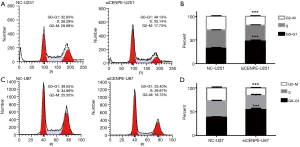
To further understand the regulatory mechanisms of CENPE, we examined changes in cell cycle after interference with CENPE. We found that cells in the G0–G1 phase increased significantly, while cells in the G2-M phase were significantly reduced in siCENPE-U251 group compared with the NC-U251 group (Figure 4A,B). U87 cells also showed the same trend (Figure 4C,D). This result indicated that the knockdown of CENPE has the greatest impact on G0–G1 transition.
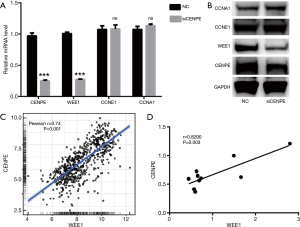
CENPE affects cell cycle by affecting the expression of WEE1
To detect what genes are downstream of CENPE, we selected genes that are thought to be involved in the cell cycle. We found that CCNA1 and CCNE1 genes did not change significantly after interference with CENPE, while the expression of WEE1 gene decreased significantly (Figure 5A,B). These results suggested that CENPE may affect the transformation of G1 phase to S phase by affecting the expression of WEE1. Then we further studied the RNA-seq data of CENPE and WEE1 in the TCGA database, and found that CENPE has a significant correlation with the expression of WEE1 (Figure 5C). At the same time, the mRNA data of the samples we collected also obtained similar results (Figure 5D).
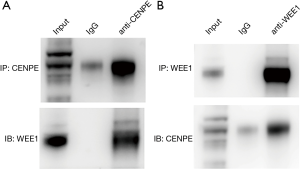
CENPE is in direct contact with WEE1
In order to further study the relationship between CENPE and WEE1, the co-IP experiments were performed. At first, we found that endogenous WEE1 proteins could be coimmunoprecipitated by CENPE antibodies in U251 cell line (Figure 6A). Then, to further determine the accuracy of this result, we performed a co-IP using the WEE1 antibody and found that the CENPE protein was also detected (Figure 6B).
Discussion
Rapid proliferation is one of the important characteristics of tumor cells, and the higher the degree of malignancy, the more proliferative ability (9,10). A better understanding of the mechanism of rapid tumor proliferation can play an important guiding role in the treatment of tumors. In addition, finding biomarkers related to rapid tumor proliferation plays an important role in the diagnosis and treatment of tumors. At present, in addition to its ability to control proliferation, many studies found that it could also be used as a biomarker for many tumors. In invasive ductal carcinoma (IDC), Studies have found that CENPE is a potentially key gene associated with IDC (11). In multiple myeloma, side population (SP) cells exhibit high CENPE expression characteristics (12). In prolactin pituitary tumors, CENPE is one of 5 genes associated with the pathological classification (13). In our study, we found that the proliferation of glioma cells was significantly inhibited after knocking down CENPE. At the same time, the expression of CENPE was not only significantly correlated with stage, but also significantly associated with IDH1 mutation. These results reflect the important characteristics of CENPE as a biomarker for tumors.
In order to ensure that the cells can be completely and normally split into two daughter cells, the cells develop three cell cycle checkpoints (G1/S phase checkpoint, G2/M phase checkpoint, spindle assembly checkpoint), allowing cell repair damaged DNA to maintain genomic integrity (14,15). In cancer cells, the G1 checkpoint mechanism of many cancer cells is defective, so, cells can proliferate rapidly and the G2 checkpoint and the spindle assembly checkpoint are far more important than normal cells (16,17). Wee1 was first identified in fission yeast, and wee1 deficiency resulted in premature mitotic entry and replication of smaller-sized yeast (18). In addition, the expression of WEE1 is also associated with the proliferation of many tumors. In glioma cells, miR-526b-3p inhibits tumor cell proliferation by inhibiting the expression of WEE1 (19). In liver cancer, upregulated DLX6-AS1 promoted cell proliferation, migration, and invasion through increasing expression of WEE1 via targeting miR-424-5p (20).
In addition, WEE1 inhibitors have been used in clinical trials. WEE1 inhibition by AZD1775 abrogates the G2-M arrest, allowing cells with a deregulated G1 into premature mitosis, resulting in mitotic catastrophe and apoptosis through unrepaired DNA damage (21). A phase I trial of AZD1775 in 25 patients with advanced solid tumors has established the maximum tolerated dose (MTD) to be 225 mg twice a day over 2.5 days per week for 2 weeks every 21 days (22). This is the first reported phase I study showed safety and efficacy of single-agent, AZD1775, especially in BRCA mutant patients (22). Furthermore, one phase II study provided clinical evidence that AZD1775 enhanced carboplatin efficacy in TP53-mutant tumors (23). Since small molecule inhibitors of WEE1 also have been used in clinical trials of glioma (24) and WEE1 is closely related to the proliferation of glioma (19). Therefore, the CENPE gene, which regulates the expression of WEE1, may also be used as a target gene for clinical treatment of glioma.
Conclusions
CENPE expression was increased in GBM tissues and was associated with clinical stage and unfavorable overall survival in glioma patients. Inhibition of CENPE expression resulted in proliferation inhibition of GBM cell. CENPE promotes GBM proliferation through WEE1 pathway and binding to WEE1.
Acknowledgments
The authors sincerely appreciate all members participated in this study.
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.40). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional/regional/national ethics committee/ethics board of Ethics Committee of Zhejiang Institute of Traditional Chinese Medicine (No. XMSC2018048). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Westphal M, Heese O, Steinbach JP, et al. A randomised, open label phase III trial with nimotuzumab, an anti-epidermal growth factor receptor monoclonal antibody in the treatment of newly diagnosed adult glioblastoma. Eur J Cancer 2015;51:522-32. [Crossref] [PubMed]
- Pinel S, Thomas N, Boura C, et al. Approaches to physical stimulation of metallic nanoparticles for glioblastoma treatment. Adv Drug Deliv Rev 2019;138:344-57. [Crossref] [PubMed]
- Testa JR, Zhou JY, Bell DW, et al. Chromosomal localization of the genes encoding the kinetochore proteins CENPE and CENPF to human chromosomes 4q24-->q25 and 1q32-->q41, respectively, by fluorescence in situ hybridization. Genomics 1994;23:691-3. [Crossref] [PubMed]
- Hou S, Li N, Zhang Q, et al. XAB2 functions in mitotic cell cycle progression via transcriptional regulation of CENPE. Cell Death Dis 2016;7:e2409. [Crossref] [PubMed]
- Liang Y, Ahmed M, Guo H, et al. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res 2017;77:5479-90. [Crossref] [PubMed]
- Dun B, Sharma A, Xu H, et al. Transcriptomic changes induced by mycophenolic acid in gastric cancer cells. Am J Transl Res 2013;6:28-42. [PubMed]
- Balamuth NJ, Wood A, Wang Q, et al. Serial transcriptome analysis and cross-species integration identifies centromere-associated protein E as a novel neuroblastoma target. Cancer Res 2010;70:2749-58. [Crossref] [PubMed]
- Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 2017;17:93-115. [Crossref] [PubMed]
- Goncalves MD, Hopkins BD, Cantley LC. Phosphatidylinositol 3-Kinase, Growth Disorders, and Cancer. N Engl J Med 2018;379:2052-62. [Crossref] [PubMed]
- Li C, Luo L, Wei S, et al. Identification of the potential crucial genes in invasive ductal carcinoma using bioinformatics analysis. Oncotarget 2017;9:6800-13. [PubMed]
- Nara M, Teshima K, Watanabe A, et al. Bortezomib reduces the tumorigenicity of multiple myeloma via downregulation of upregulated targets in clonogenic side population cells. PLoS One 2013;8:e56954. [Crossref] [PubMed]
- Raverot G, Wierinckx A, Dantony E, et al. Prognostic factors in prolactin pituitary tumors: clinical, histological, and molecular data from a series of 94 patients with a long postoperative follow-up. J Clin Endocrinol Metab 2010;95:1708-16. [Crossref] [PubMed]
- Roy D, Sheng GY, Herve S, et al. Interplay between cancer cell cycle and metabolism: Challenges, targets and therapeutic opportunities. Biomed Pharmacother 2017;89:288-96. [Crossref] [PubMed]
- Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer 2018;18:168-85. [Crossref] [PubMed]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [Crossref] [PubMed]
- Schmidt M, Rohe A, Platzer C, et al. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules 2017; [Crossref] [PubMed]
- Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature 1979;279:428-30. [Crossref] [PubMed]
- Wu M, Li X, Liu Q, et al. miR-526b-3p serves as a prognostic factor and regulates the proliferation, invasion, and migration of glioma through targeting WEE1. Cancer Manag Res 2019;11:3099-110. [Crossref] [PubMed]
- Li D, Tang X, Li M, et al. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem 2019;120:12290-9. [Crossref] [PubMed]
- Aarts M, Sharpe R, Garcia-Murillas I, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov 2012;2:524-39. [Crossref] [PubMed]
- Do K, Wilsker D, Ji J, et al. Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol 2015;33:3409-15. [Crossref] [PubMed]
- Leijen S, van Geel RM, Sonke GS, et al. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J Clin Oncol 2016;34:4354-61. [Crossref] [PubMed]
- Sanai N, Li J, Boerner J, et al. Phase 0 trial of AZD1775 in first-recurrence glioblastoma patients. Clin Cancer Res 2018;24:3820-8. [Crossref] [PubMed]


