
Clinical significance of PI3 and HLA-DOB as potential prognostic predicators for ovarian cancer
Introduction
Ovarian cancer is one of the most common malignant tumors in the female reproductive system and has the highest mortality among all gynecological tumors (1,2). Due to the absence of specific symptoms and detective tools, most ovarian cancer patients are diagnosed at advanced stages, and the 5-year survival rate remains at approximately 45% (3,4). Despite improvements in surgery and chemotherapy approaches, unfortunately, the majority of advanced patients eventually relapse and die of this disease. Moreover, the prognosis of patients remains poor, which emphasizes the importance of identifying novel biomarkers predicting patients’ outcomes.
Clinicopathological characteristics, such as age at diagnosis, tumor subtype, clinical stage, histological grade, treatment modalities, and residual disease, affect the prognosis of ovarian cancer (2,5,6). Genetic alterations, such as chromosomal rearrangement (7,8), copy number amplification (9,10), DNA methylation (11) and gene mutation (12,13), also contribute to ovarian tumorigenesis and progression. The expression levels of some genes have been discovered to have a significant relationship with clinical outcomes, and are proposed as prognostic markers (14-16).
However, there is currently a lack of systematic genome-wide screens for ovarian cancer prognostic factors. By using a self-developed pipeline, this study aims to find prognosis-related genes in The Cancer Genome Atlas (TCGA) ovarian serous cystadenocarcinoma gene expression data and to explore the potential underlying molecular mechanisms of ovarian cancer through bioinformatics methods.
Methods
Data source
Both ovarian serous cystadenocarcinoma gene expression data and clinicopathological data were downloaded from TCGA database (https://tcga.xenahubs.net/download/TCGA.OV.sampleMap). Subsequently, these data were matched by sample ID. Gene expression was measured experimentally by the Broad Institute of MIT and the Harvard University Cancer Genomic Characterization Center using Affymetrix HT Human Genome U133a microarray platform. Only primary tumor samples were kept. Finally, 12,042 genes from 562 samples were included in the data set.
Data preprocessing
Genes with the most obvious variance (upper 25%) were selected as candidate genes (n=3,011) and kept for further analysis. The expression of each gene was labeled as “low” or “high” when compared with the median expression level of that gene.
Statistical analysis
Candidate genes were subjected to Kaplan-Meier survival analysis, and OS was calculated as the number of days between the date of diagnosis and the date of death or the last follow-up, whichever came first. Statistical significance was calculated using the log-rank test. Fisher’s exact test was used to compare patients’ distribution and unknowns were excluded before the analysis. Tumor characteristics and multivariate Cox proportional hazards models were performed. Statistical tests were two-tailed and the threshold for the P value was set at <0.05. For comparisons of multiple candidate genes, the threshold for the P values was set at 1.66e-5 according to Bonferroni correction (0.05/3011).
Bioinformative analysis
In total, 542 genes that significantly correlated with PI3 or HLA-DOB (227 genes with PI3, 404 genes with HLA-DOB and 89 overlapping genes) were used for enrichment analysis, where the P value was corrected with the Benjamini and Hochberg method (BH correction).
Software and packages
R (3.3.1) (17) was used for data preprocessing and statistical analysis, and the “survival” package was used for the survival analysis. FunRich (version 3) software (18) was used for the bioinformatics analysis.
Results
Kaplan-Meier survival analysis
All 3,011 candidate genes were subjected one by one to a self-developed R program, in which Kaplan-Meier analysis carried out between different gene expression groups (“low” or “high”). Two genes showed significant associations under the Bonferroni threshold (P=1.66e-5): PI3 (P=7.99e-7) and HLA-DOB (P=7.52e-6). Patients with high PI3 levels experienced prolonged OS, while high HLA-DOB transcription showed a negative influence on OS. When samples were subdivided according to clinical stages, the effects of these two genes were observed in stage III/IV but not in stage I/II (Figure 1).
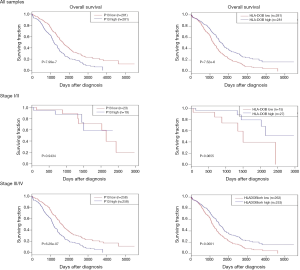
Univariate analysis between genes and clinical parameters
Then, clinicopathological characteristics were compared between the low and high expression groups, but there was no significant difference in most of them (such as age at diagnosis, clinical stage, histological grade and invasion). For PI3, the anatomic subdivision of cancer was unevenly distributed between the low and high expression groups. Low PI3 levels were significantly associated with unilateral tumors, but patients with high PI3 levels were more likely to suffer bilateral lesions. Low HLA-DOB expression was significantly related to progressive diseases, but high HLA-DOB expression was more likely as a sign of stable disease (Table 1).
Table 1
| Factors | PI3 | HLA-DOB | |||||
|---|---|---|---|---|---|---|---|
| Low (n=281) | High (n=281) | P | Low (n=281) | High (n=281) | P | ||
| Age of diagnosis (year) | 0.9267 | 0.4439 | |||||
| <50 | 59 | 55 | 51 | 63 | |||
| 50–59 | 92 | 88 | 87 | 93 | |||
| 60–69 | 65 | 69 | 71 | 63 | |||
| ≥70 | 65 | 69 | 72 | 62 | |||
| Anatomic neoplasm subdivision | 0.00816 | 0.09388 | |||||
| Bilateral | 181 | 210 | 188 | 203 | |||
| Left | 50 | 27 | 37 | 40 | |||
| Right | 35 | 29 | 40 | 24 | |||
| Unknown | 15 | 15 | 16 | 14 | |||
| Clinical stage | 0.541 | 0.09709 | |||||
| Stage I | 10 | 5 | 7 | 8 | |||
| Stage II | 13 | 14 | 8 | 19 | |||
| Stage III | 213 | 219 | 215 | 217 | |||
| Stage IV | 45 | 39 | 48 | 36 | |||
| Unknown | 0 | 4 | 3 | 1 | |||
| Histologic grade | 0.7102 | 0.3208 | |||||
| Grade 1/2 | 36 | 39 | 33 | 42 | |||
| Grade 3/4 | 240 | 234 | 240 | 234 | |||
| Unknown | 5 | 8 | 8 | 5 | |||
| Venous invasion | 0.6312 | 0.6249 | |||||
| No | 32 | 38 | 31 | 39 | |||
| Yes | 43 | 43 | 34 | 52 | |||
| Unknown | 206 | 200 | 216 | 190 | |||
| Lymphatic invasion | 0.5701 | 0.5689 | |||||
| No | 36 | 43 | 39 | 40 | |||
| Yes | 67 | 66 | 59 | 74 | |||
| Unknown | 178 | 172 | 183 | 167 | |||
| Tumor residual disease | 0.5594 | 0.04907 | |||||
| No macroscopic disease | 61 | 54 | 49 | 66 | |||
| 1–10 mm | 130 | 118 | 139 | 109 | |||
| 11–20 mm | 19 | 17 | 19 | 17 | |||
| >20 mm | 47 | 58 | 46 | 59 | |||
| Unknown | 24 | 34 | 28 | 30 | |||
| New neoplasm event | 0.3139 | 0.7704 | |||||
| Locoregional disease | 2 | 4 | 3 | 3 | |||
| Metastatic | 1 | 0 | 1 | 0 | |||
| Progression of disease | 19 | 11 | 13 | 17 | |||
| Recurrence | 133 | 127 | 133 | 127 | |||
| Unknown | 126 | 139 | 131 | 134 | |||
| Primary therapy outcome | 0.3571 | 0.04032 | |||||
| Complete remission/response | 133 | 123 | 124 | 132 | |||
| Partial remission/response | 24 | 31 | 27 | 28 | |||
| Progressive disease | 20 | 22 | 27 | 15 | |||
| Stable disease | 19 | 11 | 9 | 21 | |||
| Unknown | 85 | 94 | 94 | 85 | |||
The association between clinicopathological characteristics and gene expression levels (low or high) was analyzed by Fisher’s exact test. Unknowns were excluded before calculation.
Multivariate Cox proportional hazards analysis
Gene expression level and clinicopathological characteristics were incorporated into a multivariate Cox proportional hazards model for survival analysis. Both PI3 and HLA-DOB were revealed as independent predictors of prognosis among these factors. Compared with a lower mRNA expression level of PI3, a higher PI3 mRNA expression level had a hazard ratio (HR) of 1.84 (P=3.77e-7), while a higher mRNA expression level of HLA-DOB had a HR of 0.68 (P=0.001134). In the model, age at diagnosis (more than 70 years), tumor residual disease, new neoplasm events (such as metastasis and recurrence) and primary therapy outcomes (such as partial remission and progressive disease) were significantly associated with OS (Table 2).
Table 2
| Factors | PI3 | HLA-DOB | |||||
|---|---|---|---|---|---|---|---|
| P | Se | HR (95% CI) | P | Se | HR (95% CI) | ||
| PI3/HLA-DOB | |||||||
| Low | Reference | Reference | |||||
| High | 3.77e–07 | 0.12 | 1.84 (1.45–2.32) | 0.001134 | 0.12 | 0.68 (0.54–0.86) | |
| Age of diagnosis (year) | |||||||
| <50 | Reference | Reference | |||||
| 50–59 | 0.52317 | 0.18 | 1.12 (0.79–1.58) | 0.249306 | 0.18 | 1.23 (0.87–1.73) | |
| 60–69 | 0.14116 | 0.19 | 1.32 (0.91–1.9) | 0.089531 | 0.19 | 1.38 (0.95–1.99) | |
| ≥70 | 8.37e-06 | 0.19 | 2.31 (1.6–3.35) | 1.62e-06 | 0.19 | 2.48 (1.71–3.6) | |
| Anatomic subdivision | |||||||
| Bilateral | Reference | Reference | |||||
| Left | 0.24365 | 0.19 | 0.8 (0.55–1.17) | 0.089538 | 0.19 | 0.72 (0.5–1.05) | |
| Right | 0.43376 | 0.19 | 1.16 (0.8–1.71) | 0.884027 | 0.19 | 0.97 (0.67–1.41) | |
| Unknown | 0.83131 | 0.26 | 0.95 (0.57–1.57) | 0.565114 | 0.26 | 0.86 (0.52–1.42) | |
| Clinical stage | |||||||
| Stage I | Reference | Reference | |||||
| Stage II | 0.44687 | 0.71 | 0.58 (0.14–2.35) | 0.804682 | 0.71 | 0.84 (0.21–3.4) | |
| Stage III | 0.73549 | 0.63 | 0.81 (0.23–2.78) | 0.887218 | 0.63 | 1.09 (0.32–3.74) | |
| Stage IV | 0.75831 | 0.64 | 1.22 (0.35–4.26) | 0.513961 | 0.64 | 1.52 (0.43–5.28) | |
| Unknown | 0.55735 | 1.03 | 1.83 (0.24–13.63) | 0.320538 | 1.02 | 2.76 (0.37–20.46) | |
| Histologic grade | |||||||
| Grade1/2 | Reference | Reference | |||||
| Grade3/4 | 0.19132 | 0.17 | 1.25 (0.89–1.76) | 0.233345 | 0.17 | 1.23 (0.88–1.72) | |
| Unknown | 0.53482 | 0.42 | 0.77 (0.34–1.75) | 0.62694 | 0.41 | 1.22 (0.55–2.69) | |
| Venous invasion | |||||||
| No | Reference | Reference | |||||
| Yes | 0.23044 | 0.35 | 0.66 (0.33–1.3) | 0.329182 | 0.35 | 0.71 (0.36–1.41) | |
| Unknown | 0.72904 | 0.28 | 1.1 (0.64–1.9) | 0.953708 | 0.28 | 0.98 (0.57–1.69) | |
| Lymphatic invasion | |||||||
| No | Reference | Reference | |||||
| Yes | 0.0568 | 0.3 | 1.78 (0.98–3.22) | 0.105397 | 0.3 | 1.62 (0.9–2.91) | |
| Unknown | 0.80288 | 0.27 | 0.94 (0.56–1.58) | 0.54888 | 0.26 | 0.86 (0.51–1.43) | |
| Tumor residual disease | |||||||
| ≥20 mm | Reference | Reference | |||||
| No macroscopic disease | 0.00122 | 0.22 | 0.49 (0.32–0.76) | 0.000158 | 0.22 | 0.44 (0.29–0.67) | |
| 1–10 mm | 0.51364 | 0.15 | 0.9 (0.67–1.22) | 0.066415 | 0.15 | 0.75 (0.56–1.02) | |
| 11–20 mm | 0.29633 | 0.25 | 0.77 (0.47–1.26) | 0.175791 | 0.26 | 0.71 (0.43–1.17) | |
| Unknown | 0.0049 | 0.23 | 0.52 (0.33–0.82) | 0.003007 | 0.24 | 0.5 (0.31–0.79) | |
| New neoplasm event | |||||||
| Locoregional disease | Reference | Reference | |||||
| Metastatic | 0.01282 | 1.17 | 18.52 (1.86–184.46) | 0.04181 | 1.17 | 10.75 (1.09–105.78) | |
| Progression of disease | 0.04482 | 0.57 | 3.12 (1.03–9.51) | 0.042067 | 0.57 | 3.17 (1.04–9.66) | |
| Recurrence | 0.00982 | 0.55 | 4.13 (1.41–12.1) | 0.008815 | 0.55 | 4.26 (1.44–12.59) | |
| Unknown | 0.1781 | 0.56 | 2.12 (0.71–6.3) | 0.115512 | 0.56 | 2.41 (0.81–7.22) | |
| Primary therapy outcome | |||||||
| Complete remission | Reference | Reference | |||||
| Partial remission | 1.14e-09 | 0.19 | 3.17 (2.19–4.6) | 3.94e-11 | 0.19 | 3.53 (2.43–5.13) | |
| Progressive disease | 1.84e-08 | 0.23 | 3.73 (2.36–5.89) | 6.20e-09 | 0.23 | 3.87 (2.45–6.12) | |
| Stable disease | 5.91e-06 | 0.27 | 3.32 (1.98–5.59) | 7.19e-05 | 0.26 | 2.85 (1.7–4.79) | |
| Unknown | 0.00546 | 0.19 | 1.7 (1.17–2.47) | 0.002214 | 0.19 | 1.79 (1.23–2.59) | |
Influence of highlighted gene (PI3 or HLA-DOB) and clinicopathologic factors on overall survival were calculated with multivariate Cox proportional hazards analysis. HR, hazard ratio; CI, confidence interval; Se, standard error.
Coexpression and enrichment analyses
A coexpression analysis was performed to explore the potential mechanisms of PI3 and HLA-DOB in ovarian cancer prognosis. The 20 genes that most significantly correlated with ovarian cancer prognosis are listed in Table 3, where SOD2 (superoxide dismutase 2) (with PI3) and TAP1 (transporter 1, ATP binding cassette subfamily B member) (with HLA-DOB) ranked first. Pathway and transcription factor analyses were carried out on genes that were coexpressed with these two genes. The most remarkable pathways are listed in Table 4, where “immune system” ranked first. Meanwhile, IRF1 (P=1.16e-15) and SPI1 (P=2.03e-6) were the most significant molecules in the transcription factor analysis, as shown in Table 5.
Table 3
| Order | PI3 | HLA-DOB | |||||
|---|---|---|---|---|---|---|---|
| Gene | P | r | Gene | P | r | ||
| 1 | SOD2 | 2.03e-34 | 0.484 | TAP1 | 1.66e-54 | 0.592 | |
| 2 | SLPI | 2.34e-33 | 0.478 | PSMB9 | 4.07e-51 | 0.577 | |
| 3 | CXCL8 | 3.01e-30 | 0.456 | PSMB8 | 1.88e-49 | 0.569 | |
| 4 | CXCL1 | 3.35e-30 | 0.456 | BTN3A3 | 9.86e-46 | 0.550 | |
| 5 | S100A8 | 6.72e-27 | 0.432 | CYCSP5 | 3.39e-45 | 0.547 | |
| 6 | CCL20 | 4.76e-26 | 0.425 | HLA-DMA | 1.33e-44 | 0.544 | |
| 7 | S100A9 | 8.97e-26 | 0.423 | CXCL11 | 5.26e-41 | 0.524 | |
| 8 | LCN2 | 2.21e-23 | 0.403 | HLA-F | 3.93e-40 | 0.519 | |
| 9 | C1S | 6.51e-22 | 0.390 | APOL3 | 1.71e-39 | 0.516 | |
| 10 | ICAM1 | 1.24e-20 | 0.379 | HLA-E | 2.82e-38 | 0.508 | |
| 11 | PTX3 | 4.06e-20 | 0.374 | HLA-DMB | 6.13e-38 | 0.506 | |
| 12 | RARRES1 | 3.77e-19 | 0.365 | UBD | 1.12e-37 | 0.505 | |
| 13 | NFKBIA | 4.42e-19 | 0.364 | HLA-DRB1 | 4.59e-37 | 0.501 | |
| 14 | PDZK1IP1 | 5.30e-19 | 0.364 | IRF1 | 1.46e-36 | 0.498 | |
| 15 | PLAUR | 4.53e-18 | 0.354 | CD38 | 7.09e-35 | 0.487 | |
| 16 | CXCL5 | 2.19e-17 | 0.347 | HLA-DPB1 | 7.95e-35 | 0.487 | |
| 17 | HP | 1.04e-16 | 0.340 | HLA-DRA | 8.00e-35 | 0.487 | |
| 18 | CFB | 1.36e-16 | 0.339 | CD74 | 1.25e-34 | 0.486 | |
| 19 | BCL2A1 | 1.85e-16 | 0.338 | TMEM140 | 6.62e-34 | 0.481 | |
| 20 | TNFAIP6 | 1.08e-15 | 0.329 | HLA-DPA1 | 8.97e-34 | 0.480 | |
Correlations between highlighted gene (PI3 or HLA-DOB) and other genes were calculated and the most significant genes with them were listed, respectively.
Table 4
| Biological pathway | No. of genes | Fold enrichment | P value (BH corrected) |
|---|---|---|---|
| Immune system | 79 | 5.460688 | 1.15e-32 |
| Cytokine Signaling in Immune system | 44 | 8.226512 | 6.66e-25 |
| Interferon signaling | 33 | 12.1512 | 2.11e-24 |
| Interferon alpha/beta signaling | 27 | 12.65382 | 2.37e-20 |
| Interferon gamma signaling | 18 | 13.8219 | 5.08e-14 |
| Epithelial-to-mesenchymal transition | 28 | 5.46213 | 6.95e-11 |
| Integrin family cell surface interactions | 86 | 2.25677 | 1.67e-10 |
| Innate Immune system | 26 | 5.127547 | 1.66e-09 |
| TRAIL signaling pathway | 81 | 2.205787 | 2.17e-09 |
Table 5
| Transcription factor | No. of genes | Fold enrichment | P value (BH corrected) |
|---|---|---|---|
| IRF1 | 71 | 3.158911 | 1.16e-15 |
| SPI1 | 52 | 2.305161 | 2.03e-06 |
| NFIC | 83 | 1.468055 | 0.010919 |
| ELF1 | 31 | 1.912417 | 0.010919 |
| FOS | 72 | 1.48117 | 0.010919 |
| FOSB | 72 | 1.48117 | 0.010919 |
| JUN | 72 | 1.48117 | 0.010919 |
| JUNB | 72 | 1.48117 | 0.010919 |
| JUND | 72 | 1.48117 | 0.010919 |
Discussion
The prognosis of cancer patients can be influenced by many regulatory molecules, so it is challenging to search for significant genes associated with outcomes. The range for candidate genes should be extended, and many clinical features (including age, stage, histopathology, etc.) should be incorporated into the model. Thus, it cannot only improve the performance of screening but also help to explain the possible mechanisms involved. In our study, genome-wide screening was performed on 12,042 genes, and our selections were tested in a model containing extensive factors, which improved the reliability and interpretability of the results.
PI3, which encodes an elastase-specific inhibitor, functions as an antimicrobial peptide against bacteria, fungi and other inflammatory pathologies (19-21). Adam Clauss et al. reported that the PI3 protein was overexpressed in serous ovarian carcinomas and showed a significant association with poor OS in 2010 (22). Further analysis confirmed the relationship between PI3 overexpression and the short survival time of ovarian tumor patients (23,24). In addition, a high level of PI3 was also related to the poor outcomes of breast cancer patients (23,25), cutaneous graft-versus-host disease (26) and hematopoietic cell transplantation (27). Moreover, the level of PI3 was related to breast cancer (25) and esophagus squamous cell carcinomas (28). In ovarian cancer, the PI3 protein can promote cell proliferation (24) and decrease epithelial ovarian carcinoma (EOC) cell sensitivity to genotoxic agents (29). However, little is known about the function of PI3 in tumor progression, and its importance has not been completely assessed from a genomic perspective.
HLA-DOB is one of the two components (the beta chain) belonging to HLA-DO, a human leucocyte antigen (HLA) class II heterodimer. HLA-DO controls HLA-DM-mediated peptide loading onto MHC class II molecules and functions as a modulator of antigen presentation (30). Polymorphisms in HLA-DOB have been identified to have significant associations with several pathology processes, such as HCV infection and viral clearance (31,32), immune control of HIV-1 infection (33) and the poor prognosis of advanced-stage non-small cell lung cancer (NSCLC) (34). To date, the relationship between HLA-DOB and ovarian cancer has rarely been reported.
Our study indicated that the level of PI3 was an independent predictor for the prognosis of ovarian cancer patients, but there are still some inconsistencies with former reports, which the association between PI3 and OS was observed in stage I/II but not in stage III/IV (23). A possible explanation for this discrepancy is that the previous study used immunohistochemistry (IHC) to assess the PI3 protein, but we focused on the level of mRNA. Moreover, as most ovarian cancers are diagnosed at an advanced stage, the sample sizes in stage I/II were relatively small, which may have reduced the reliability of the results. Larger sample sizes are needed for further study in the future.
According to Adam Clauss and colleagues, no significant differences were found in the distribution of clinicopathological characteristics (age, debulking, stage, and platinum sensitivity or resistance) between the two PI3-expression level groups (22). However, Caruso reported that patients in the PI3-positive group had a higher proportion of advanced FIGO stages (III/IV) (23). In our study, almost all clinicopathological characteristics, including clinical stage, were evenly distributed in the PI3-low and high groups except for anatomic subdivision, but our PI3-grouping was based mainly on mRNA expression instead of IHC staining (Table 1). An uneven distribution of HLA-DOB was found in the primary therapy outcome. The majority of samples with “stable disease” showed high levels, while more samples with “progressive disease” were sorted into the low group, implying that HLA-DOB may affect the tumor’s response to treatment.
In the Cox proportional hazards test (Table 2), more variables were taken into account than those described in other studies (22-24). Both PI3 and HLA-BOD showed a significant influence on OS together with venous/lymphatic invasion, tumor residual disease, new neoplasm events and primary therapy outcomes, demonstrating that these two genes have good predictive value for ovarian cancer prognosis. An age at diagnosis of over 70 years was identified as a risk factor, while “no macroscopic disease” was identified as a protective factor (relative to their respective references). Among all factors, new neoplasm events (metastatic/progression of disease/recurrence) and primary therapy outcomes (partial remission/progressive disease/stable disease) had a decisive impact on OS. All of these results agreed with the consensus and confirmed the validity of our model.
Among the top 20 genes correlated with PI3, S100A8 (S100 calcium binding protein A8), S100A9 (S100 calcium binding protein A9) and NFKBIA (AFKB inhibitor alpha) participate in endogenous TLR (Toll-like receptor) signaling, while SOD2, CXCL8 (C-X-C motif chemokine ligand 8), ICAM1 (intercellular adhesion molecule 1), NFKBIA (NFKB inhibitor alpha), PLAUR (plasminogen activator, urokinase receptor) and BCL2A1 (BCL2 related protein A1) are involved in multiple biological pathways, such as PI3K-mTOR (phosphatidylinositol 3 kinase-mammalian target of rapamycin) and EGF (epidermal growth factor) receptor signaling. Among the top 20 genes correlated with HLA-DOB, many are related to immune system regulation such as antigen processing/presentation, the interferon pathway and cytokine signaling. The close relationship between the correlated genes and immunomodulation is also shown in Table 4. IRF1, which is a transcriptional regulator involved in both innate and acquired immune responses was revealed in the transcription factor analysis. IRF1 expression can be induced by cisplatin and attenuates drug sensitivity in ovarian cancer cells (35). It has also been identified as an independent predictor of prognosis in high-grade serous ovarian carcinoma (HGSOC) (36). SPI1 is an ETS-domain transcription factor that activates gene expression during myeloid and B-lymphoid cell development (37), but the role of SPI1 in ovarian cancer is not clear.
Our results were obtained from statistical and bioinformation analyses and further experimental and clinical studies are warranted to verify these findings.
Acknowledgments
We acknowledge the contributions of the TCGA Research Network.
Funding: This study was supported by
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovairan cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Matulonis UA, Sood AK, Fallowfield L, et al. Ovarian cancer. Nat Rev Dis Primers 2016;2:16061. [Crossref] [PubMed]
- Escayola C, Ferron G, Romeo M, et al. The impact of pleural disease on the management of advanced ovarian cancer. Gynecol Oncol 2015;138:216-20. [Crossref] [PubMed]
- Elzakkers JCJ, van der Aa MA, van Altena AM, et al. Further insights into the role of tumour characteristics in survival of young women with epithelial ovarian cancer. Gynecol Oncol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Firat Cuylan Z, Karabuk E, Oz M, et al. Comparison of stage III mucinous and serous ovarian cancer: a case-control study. J Ovarian Res 2018;11:91. [Crossref] [PubMed]
- Hillman RT, Chisholm GB, Lu KH, et al. Genomic Rearrangement Signatures and Clinical Outcomes in High-Grade SerousOvarian Cancer. J Natl Cancer Inst 2018;110:265-72. [Crossref] [PubMed]
- da Costa AABA, do Canto LM, Larsen SJ, et al. Genomic profiling in ovarian cancer retreated with platinum based chemotherapy presented homologous recombination deficiency and copy number imbalances of CCNE1 and RB1 genes. BMC Cancer 2019;19:422. [Crossref] [PubMed]
- Morikawa A, Hayashi T, Kobayashi M, et al. Somatic copy number alterations have prognostic impact in patients with ovarianclear cell carcinoma. Oncol Rep 2018;40:309-18. [PubMed]
- Stronach EA, Paul J, Timms KM, et al. Biomarker assessment of HR deficiency, tumor BRCA1/2 mutations, and CCNE1 copy number in ovarian cancer: associations with clinical outcome following patinum monotherapy. Mol Cancer Res 2018;16:1103-11. [Crossref] [PubMed]
- Bodelon C, Killian JK, Sampson JN, et al. Molecular classification of epithelial ovarian cancer based on methylation profiling: evidence for survival heterogeneity. Clin Cancer Res 2019;25:5937-46. [Crossref] [PubMed]
- Yang SYC, Lheureux S, Karakasis K, et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med 2018;10:81. [Crossref] [PubMed]
- Suszynska M, Klonowska K, Jasinska AJ, et al. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes - Providing evidence of cancer predisposition genes. Gynecol Oncol 2019;153:452-62. [Crossref] [PubMed]
- Sun X, Wang S, Li Q. Comprehensive analysis of expression and prognostic value of Sirtuins in ovarian cancer. Front Genet 2019;10:879. [Crossref] [PubMed]
- Hinchcliff E, Paquette C, Roszik J, et al. Lymphocyte-specific kinase expression is a prognostic indicator in ovarian cancerand correlates with a prominent B cell transcriptional signature. Cancer Immunol Immunother 2019;68:1515-26. [Crossref] [PubMed]
- Tsibulak I, Wieser V, Degasper C, et al. BRCA1 and BRCA2 mRNA-expression prove to be of clinical impact in ovarian cancer. Br J Cancer 2018;119:683-92. [Crossref] [PubMed]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2016.
- Pathan M, Keerthikumar S, Ang CS, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597-601. [Crossref] [PubMed]
- Wang Z, Chen F, Zhai R, et al. Plasma neutrophil elastase and elafin imbalance is associated with acute respiratory distress syndrome (ARDS) development. PloS One 2009;4:e4380. [Crossref] [PubMed]
- Kerrin A, Weldon S, Chung AH, et al. Proteolytic cleavage of elafin by 20S proteasome may contribute to inflammation in acute lung injury. Thorax 2013;68:315-21. [Crossref] [PubMed]
- Motta JP, Bermudez-Humaran LG, Deraison C, et al. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci Transl Med 2012;4:158ra144. [Crossref] [PubMed]
- Clauss A, Ng V, Liu J, et al. Overexpression of elafin in ovarian carcinoma is driven by genomic gains and activation of the nuclear factor kappaB pathway and is associated with poor overall survival. Neoplasia 2010;12:161-72. [Crossref] [PubMed]
- Caruso JA, Karakas C, Zhang J, et al. Elafin is downregulated during breast and ovarian tumorigenesis but its residual expression predicts recurrence. Breast Cancer Res 2014;16:3417. [Crossref] [PubMed]
- Labidi-Galy SI, Clauss A, Ng V, et al. Elafin drives poor outcome in high-grade serous ovarian cancers and basal-like breast tumors. Oncogene 2015;34:373-83. [Crossref] [PubMed]
- Hunt KK, Wingate H, Yokota T, et al. Elafin, an inhibitor of elastase, is a prognostic indicator in breast cancer. Breast Cancer Res 2013;15:R3. [Crossref] [PubMed]
- Brüggen MC, Petzelbauer P, Greinix H, et al. Epidermal elafin expression is an indicator of poor prognosis in cutaneous graft-versus-host disease. J Invest Dermatol 2015;135:999-1006. [Crossref] [PubMed]
- Hingorani S, Finn LS, Pao E, et al. Urinary elafin and kidney injury in hematopoietic cell transplant recipients. Clin J Am Soc Nephrol 2015;10:12-20. [Crossref] [PubMed]
- Yamamoto S, Egami H, Kurizaki T, et al. Immunohistochemical expression of SKALP/elafin in squamous cell carcinoma of the oesophagus. Br J Cancer 1997;76:1081-86. [Crossref] [PubMed]
- Wei H, Hellstrom KE, Hellstrom I. Elafin selectively regulates the sensitivity of ovarian cancer cells to genotoxic drug-induced apoptosis. Gynecol Oncol 2012;125:727-33. [Crossref] [PubMed]
- Liljedahl M, Kuwana T, Fung-Leung WP, et al. HLA-DO is a lysosomal resident which requires association with HLA-DM for efficient intracellular transport. EMBO J 1996;15:4817-24. [Crossref] [PubMed]
- Huang P, Zhang Y, Lu X, et al. Association of polymorphisms in HLA antigen presentation-related genes with the outcomes of HCV infection. PloS One 2015;10:e0123513. [Crossref] [PubMed]
- Huang P, Dong L, Lu X, et al. Genetic variants in antigen presentation-related genes influence susceptibility to hepatitis C virus and viral clearance: a case control study. BMC Infect Dis 2014;14:716. [Crossref] [PubMed]
- Prentice HA, Pajewski NM, He D, et al. Host genetics and immune control of HIV-1 infection: fine mapping for the extended human MHC region in an African cohort. Genes Immun 2014;15:275-81. [Crossref] [PubMed]
- Pu X, Hildebrandt MA, Lu C, et al. Inflammation-related genetic variations and survival in patients with advanced non-small cell lung cancer receiving first-line chemotherapy. Clin Pharmacol Ther 2014;96:360-9. [Crossref] [PubMed]
- Pavan S, Olivero M, Cora D, et al. IRF-1 expression is induced by cisplatin in ovarian cancer cells and limits drug effectiveness. Eur J Cancer 2013;49:964-73. [Crossref] [PubMed]
- Cohen S, Mosig R, Moshier E, et al. Interferon regulatory factor 1 is an independent predictor of platinum resistance and survival in high-grade serous ovarian carcinoma. Gynecol Oncol 2014;134:591-8. [Crossref] [PubMed]
- Verbiest T, Bouffler S, Nutt SL, et al. PU.1 downregulation in murine radiation-induced acute myeloid leukaemia (AML): from molecular mechanism to human AML. Carcinogenesis 2015;36:413-9. [Crossref] [PubMed]


