
SRC-1 and Twist1 are prognostic indicators of liver cancer and are associated with cell viability, invasion, migration and epithelial-mesenchymal transformation of hepatocellular carcinoma cells
Introduction
Hepatocellular carcinoma (HCC), the fifth and eighth leading cause of cancer in men and women, respectively, is the second leading cause of worldwide cancer-related deaths, and its incidence rate is still increasing (1,2). Despite the development of treatment strategies such as liver transplantation and hepatic resection, the 5-year survival rate of HCC is thought to be the second lowest among cancers, with less than 16% of patients among all tumor types surviving to 5 years, due to cancer metastasis and recurrence and a lack of effective therapeutic agents (3). Since most HCC patients are in advanced stages when they are diagnosed, the early, accurate and effective diagnosis of HCC would be very helpful for treatment and could improve survival rates (4). However, HCC patients are not diagnosed until they have reached the intermediate or advanced stages of disease. Thus, it is very important to further understand the molecular mechanisms, especially the oncogenic mechanisms, and to develop better diagnostic methods for HCC in the future.
Studies show that many genes are abnormally expressed in cancer patients. Among these oncogenes, Twist1, which encodes a transcription factor that is a member of a protein family involved in the regulation of organogenesis, is reported to play a role in many cancers, such as endometrial cancer (5), breast cancer (6), and liver cancer (7). However, despite these reports, clinical evidence for the relationship between Twist1 and liver cancer is still lacking.
Recently, a study showed that the expression of Twist1 and another oncogene, SRC-1, which is mainly abnormally expressed in breast cancer (8) and prostate cancer (9), is positively correlated with poor prognosis in human breast cancer (10). SRC-1 is also thought to regulate the expression of Twist during the epithelial-mesenchymal transition (11). However, to the best of our knowledge, no study has focused on the role of SRC-1 in liver cancer and its relationship with Twist1.
In the present study, we aimed to analyze the roles of SRC-1 and Twist1 in liver cancer and explore the correlation of their expression with the prognosis of patients with liver cancer. This study might give evidence for the clinical usefulness of SRC-1 and Twist1 as prognostic factors and might provide new potential tools for both the diagnosis of HCC and the treatment of HCC patients.
Methods
Patients
The present study included a total of 70 patients who underwent liver transplantation or hepatic resection surgeries in our hospital from May 2016 to December 2017. All patients were initially diagnosed with HCC, which was confirmed by pathological study. Patients who received any chemotherapy before the surgery were excluded. Patients’ characteristics, including age, sex, tumor stage, tumor-node-metastasis (TNM) grade and cirrhosis condition, were collected. Alpha-fetoprotein (AFP) and hepatitis B virus (HBV) data were also examined. All of the patients were followed up by a clinical interview or phone call. The total follow-up period was 12–60 months. For survival analysis, the overall survival (OS) time was calculated from the date of diagnosis to the date of death or the date of last contact, while the survival condition was defined as the duration from the date of diagnosis to the date of documented disease progression. The present study was approved by the ethics committee of Guizhou Provincial People’s Hospital [Approval No. EC Review (Animal) 2019-008]. Informed consent was obtained from all patients.
Immunohistochemistry (IHC)
The tissues were immediately collected after resection and stored at –20 °C before use. Briefly, all tissues were fixed with 10% formalin, embedded in paraffin and sectioned. Samples were then stained with hematoxylin and eosin (HE), immersed in 3% H2O2 and incubated with primary antibodies (anti-SRC-1 and anti-Twist1, R&D, Cambridge, MA, USA) at 4 °C overnight. Tissues were then incubated with the corresponding secondary antibody (peroxidase-conjugated rat anti-goat IgG, 1:200, ZSBio, China) at 37 °C for 30 min and stained with diaminobenzidine (DAB). Samples of HCC with high SRC-1 or Twist1 expression served as positive controls, and sections treated with species-matched, normal non-immune IgG were used as negative controls (NCs). The visualization of tan granules in the cytoplasm was considered positive. The Allred scoring system was used for the pathologic scoring (12): the sum of staining intensity 0 (no staining), 1 (weak staining), 2 (moderate staining, tan) and 3 (strong staining, dark brown) and the percentage scores of the stained area, 0 (none); 1 (2–25%); 2 (26–50%); 3 (51–75%); and 4 (>75%). The product of the staining intensity × the percentage of the stained area was regarded as the final IHC score (0–4, negative; >4, positive).
Cell culture and transfection
HCC cell lines HCCC9810, BEL-7402, Huh-7, SMMC-7721, HEPG2 and the immortalized normal liver cell line THLE-3 were all purchased from ATCC (Manassas, VA, USA). Briefly, cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% Gibco® fetal bovine serum (FBS) and 100 µg/mL penicillin-streptomycin (Sigma-Aldrich Co., USA) and were maintained at 37 °C with 5% CO2. Sh-RNA for SRC-1 (sh-SRC-1) and the NC were purchased from GeneChem Corp., Shanghai, China. Cells were transfected with sh-SRC-1 or sh-NC (5 nM) using the Lipo6000 reagent (Beyotime Biotechnology, China) according to the manufacturer’s instructions. After 48 h of transfection, the transfected cells were harvested and analyzed by quantitative real-time polymerase chain reaction (qRT-PCR).
MTT assays
Cell viability was evaluated using the MTT assay. Briefly, cells with a density of 4×104 were cultured in 96-well plates for 48 h, and 10 µL MTT solution (5 mg/mL) was added. The cells were further cultured for 4 h at 37 °C with 5% CO2. Then, the MTT solution was removed, and the supernatant was subsequently replaced with 180 µL DMSO. The optical density (OD) was evaluated at 490 nm.
In vitro cell invasion and migration assays
Both cell invasion and migration were determined by transwell assay. Briefly, for a migration assay cell suspension (2.5–5.3×104 cells/mL) was plated in the top chamber with a non-coated membrane (BD Biosciences). For the invasion assay, cells (1.25×105) were plated in the top chamber with a Matrigel-coated membrane (BD Biosciences). In both assays, cells were plated in serum-free media and were incubated for 24 h. The cells were then stained with 0.1% crystal violet, counted and photographed. All experiments were conducted in triplicate.
Western blotting analysis
The extracted protein samples were separated by SDS-PAGE and transferred to PVDF membranes. Samples were then blocked with 5% non-fat milk at room temperature for 1 h and subsequently probed with specific primary antibodies (Abcam, USA) against SRC-1 (anti-SRC-1 antibody, ab5407, 1/2,000), Twist (anti-Twist antibody, ab50581, 1/1,000), N-cadherin (anti-N-cadherin antibody, ab18203, 1 µg/mL), vimentin (anti-vimentin antibody, ab8978, 1/1,000) and E-cadherin (anti-E-cadherin antibody, ab1416, 1/50) at 4 °C overnight; subsequently, they were incubated with a corresponding secondary antibody at 37 °C for 45 min. The target bands were then visualized using enhanced chemiluminescence (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control.
Statistical analysis
A chi-square test was used to compare the quantitative factors and their rates, and comparisons in groups of continuous data were performed using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Spearman analysis was used to determine the correlation of SRC-1 and Twist1 expression. A Kaplan-Meier curve was performed for the survival analysis and to assess the relationship between the levels of SRC-1 and Twist1 and the 5-year survival of patients. Risk factors for 5-year survival rates were analyzed using Cox univariate and multivariate analysis. A P value less than 0.05 was considered statistically significant. All calculations were made using GraphPad Prism 6.0.
Results
The expression of SRC-1 and Twist1 was positively correlated in HCC patients
Expression of SRC-1 and Twist1 was determined by IHC analysis, and the correlation was calculated using Spearman analysis. As shown in Figure 1 and Table 1, the expression of SRC-1 was positive in 40 (57.1%) patients, and Twist1 was positive in 46 (65.7%) patients. Subsequent Spearman analysis showed that the expression of SRC-1 and Twist1 was positively correlated in HCC patients (P<0.05).

Table 1
| Positive/negative | SRC-1 | P value | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Twist1 | ||||
| Positive | 34 | 12 | 46 | – |
| Negative | 6 | 18 | 24 | 0.0009 |
| Total | 40 | 30 | 70 | – |
The expression of SRC-1 and Twist1 was correlated with clinical outcomes of HCC patients
To further investigate the role of SRC-1 and Twist1 in HCC, the relationship of SRC-1 and Twist1 expression with the clinical outcomes of HCC patients was studied. The results showed that the expression of SRC-1 was significantly different in patients with different tumor stages, TNM grades, and states of liver cirrhosis (P<0.05). The patients without SRC-1 had smaller tumor diameters, lower rates of TNM III–IV and lower percentages of liver cirrhosis (P<0.05, Table 2). Meanwhile, a lower rate of TNM III–IV and a lower percentage of liver cirrhosis were observed in patients who were negative for Twist1 (P<0.05). However, no significant difference was found in other indexes. These results indicate that the expression of SRC-1 and Twist1 is correlated with the clinical outcomes of HCC patients.
Table 2
| Variables | Case | SRC-1 expression | Twist1 expression | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P | Positive | Negative | P | |||
| Age | ||||||||
| >50 | 44 | 26 | 18 | 0.668 | 27 | 17 | 0.318 | |
| ≤50 | 26 | 14 | 12 | 19 | 7 | |||
| Tumor stage | ||||||||
| pT1/2, ≤5 cm | 39 | 16 | 23 | 0.00224* | 21 | 18 | 0.0409* | |
| pT3/4, >5 cm | 31 | 24 | 7 | 24 | 7 | |||
| Grade | ||||||||
| I/II | 33 | 12 | 21 | 0.00037* | 15 | 18 | 0.00074* | |
| III/IV | 37 | 29 | 8 | 31 | 6 | |||
| Gender | ||||||||
| Male | 64 | 37 | 27 | 0.711 | 44 | 20 | 0.082 | |
| Female | 6 | 3 | 3 | 2 | 4 | |||
| HBV | ||||||||
| Positive | 59 | 36 | 23 | 0.321 | 40 | 19 | 0.275 | |
| Negative | 10 | 4 | 6 | 5 | 5 | |||
| Unknow | 1 | 1 | 0 | 0 | 1 | |||
| AFP (ng/mL) | ||||||||
| ≤400 | 31 | 18 | 13 | 0.890 | 20 | 11 | 0.852 | |
| >400 | 39 | 22 | 17 | 26 | 13 | |||
| Unknow | 0 | 0 | 0 | 0 | 0 | |||
| Liver cirrhosis | ||||||||
| Yes | 53 | 36 | 17 | 0.0013* | 39 | 14 | 0.0143* | |
| No | 17 | 4 | 13 | 7 | 10 | |||
*, statistically significant. HBV, hepatitis B virus; AFP, alpha-fetoprotein.
The positive expression of SRC-1 and Twist1 was correlated with OS and disease-free survival time of HCC patients
Then, the relationship between the expression of SRC-1 and Twist1 and the survival conditions of HCC patients was studied. As shown in Figure 2A,B, the expression of both SRC-1 and Twist1 was significantly associated with the OS of HCC patients (P<0.05). Then, we combined the two factors and divided the patients into 4 groups: (I) SRC-1 (+) and Twist1 (+); (II) SRC-1 (+) and Twist1 (–); (III) SRC-1 (–) and Twist1 (+); (IV) SRC-1 (–) and Twist1 (–). The results showed that patients in the SRC-1 (+) and Twist1 (+) group had the lowest OS, while patients in the SRC-1 (–) and Twist1 (–) group had the highest OS; these results suggest that the expression levels of both SRC-1 and Twist1 were correlated with the OS and disease-free survival time of HCC patients (Figure 2C).
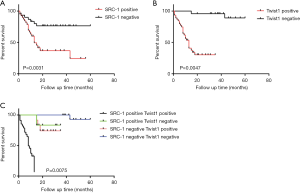
Logistic regression analysis showed the influence of different factors on the prognosis of HCC patients
The risk factors influencing the prognosis of HCC patients were then investigated using a logistic regression analysis. Cox univariate analysis showed that tumor stage, TNM grade, SRC-1 positive, Twist1 positive and SRC-1/Twist1 double positive were all significantly associated with the OS of the enrolled 70 HCC patients (P<0.05) (Table 3). Additional multivariate analysis showed that SRC-1/Twist1 double-positive and SRC-1, tumor stage, Twist1 were independent risk factors for the OS.
Table 3
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | ||
| Age | 1 | – | |||||
| >50 vs. ≤50 | 0.439 | 0.165–1.12 | 0.789 | – | – | – | |
| Stage | 1 | 1 | |||||
| T1/2 vs. T3/4 | 5.428 | 2.213–12.361 | 0.0000* | 4.845 | 2.109–11.789 | 0.000* | |
| Grade | 1 | – | |||||
| I/II vs. III/IV | 0.318 | 0.167–0.109 | 0.026* | – | – | – | |
| Gender | 1 | – | |||||
| Male vs. female | 2.058 | 0.998–4.225 | 0.055 | – | – | – | |
| HBV | 1 | – | |||||
| Negative vs. positive | 1.295 | 0.601–2.712 | 0.613 | – | – | – | |
| AFP (ng/mL) | 1 | – | |||||
| ≤400 vs. >400 | 0.612 | 0.237–1.776 | 0.315 | – | – | – | |
| Liver cirrhosis | 1 | – | |||||
| No vs. yes | 1.150 | 0.483–2.789 | 0.926 | – | – | – | |
| SRC-1 | 1 | 1 | |||||
| Negative vs. positive | 3.257 | 1.326–9.128 | 0.0056* | 3.167 | 1.246–7.935 | 0.020* | |
| Twist1 | 1 | 1 | |||||
| Negative vs. positive | 2.578 | 1.254–7.457 | 0.031* | 3.481 | 1.465–10.274 | 0.0061* | |
| SRC-1/Twist1 | 1 | – | |||||
| All others vs. SRC-1+/Twist1+ | 3.621 | 1.673–8.191 | 0.0027* | 1.679 | 1.052–3.349 | 0.004* | |
*, statistically significant. OS, overall survival; HBV, hepatitis B virus; AFP, alpha-fetoprotein; CI, confidence interval.
Inhibition of SRC-1 suppressed proliferation, invasion, migration and epithelial-mesenchymal transformation (EMT) of HCC cells
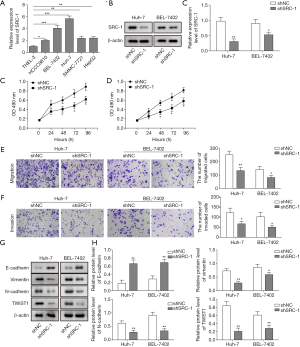
Finally, we investigated the effects of SRC-1 knockdown on HCC cell lines. As shown in Figure 3A, SRC-1 was significantly upregulated in all HCC cells compared with the normal THLE-3 cells (P<0.05). Then, SRC-1 was successfully silenced by shRNA treatment in Huh-7 and BEL-7402 cells (sh-SRC-1 group, Figure 3B). The MTT assay showed that inhibition of SRC-1 remarkably decreased cell viability in both Huh-7 and BEL-7402 cells (P<0.05, Figure 3C,D). Similarly, when SRC-1 was silenced, both cell invasion and migration were dramatically suppressed (P<0.05, Figure 3E,F). In addition, inhibition of SRC-1 also induced significant downregulation of N-cadherin and vimentin and upregulation of E-cadherin (P<0.05, Figure 3G,H). The expression of Twist1, which is a transcription factor involved in EMT progression, was also dramatically reduced following the silencing of SRC-1 (P<0.05). All these results indicated that silencing SRC-1 remarkably inhibited cell proliferation, invasion and migration as well as EMT in HCC cells, and the effects might be related to the downregulation of Twist.
Discussion
With approximately 700,000 deaths reported annually, HCC is thought to be the second leading cause of worldwide cancer-related deaths (13). It is widely accepted that early diagnosis would be beneficial for HCC treatment and would enhance the survival rates of patients (14). However, despite tremendous efforts devoted to diagnosing and managing HCC, the early diagnosis of HCC is still a major challenge, and the detailed molecular pathophysiology of HCC remains unclear. In recent decades, with the development of tumor genetics, numerous oncogenes have been found to be associated with tumorigenesis. Both Twist1 and SRC-1 are reported to be associated with the development of many cancers. Recently, Xu et al. showed that SRC-1 and Twist1 were positively correlated with a poor prognosis in human breast cancer (10). However, to the best of our knowledge, no study has focused on the role of SRC-1 in liver cancer or the relationship between SRC-1 and Twist1 in that context. In the present study, we demonstrated for the first time that both Twist1 expression and SRC-1 expression were correlated with the clinical outcomes and prognoses of HCC patients and that the silencing of SRC-1 suppressed the cell viability, invasion, migration and EMT of HCC cells, possibly through the inhibition of Twist.
Twist1 and SRC-1 were reported to be associated in many several cancers. Qin et al. showed that SRC-1 could regulate Twist expression in breast cancer and could promote cancer metastasis (11). An in vitro study also showed that Src could mediate Twist signaling and promote tumor invasion by breast tumor cells (15). In the present study, we also found that the expression of Twist1 and SRC-1 was positively correlated in HCC patients, which was consistent with previous studies. In addition, we found that inhibition of SRC-1 could suppress cancer development in HCC cells and that Twist was involved in the process, further confirming that SRC-1 could facilitate HCC and might be a promising biomarker as well as a treatment target.
There are many risk factors associated with HCC. Among the factors, HBV and AFP are 2 that are widely known. Studies have shown that chronic HBV infection is one of the major risk factors for HCC (16). Chronic HBV infection accounts for approximately 50% of total HCC cases and virtually all childhood cases of HCC (17). Similarly, AFP is another new biomarker for HCC that is helpful for monitoring HCC progression and selecting the best therapy, and it can be used as a biomarker for HCC recurrence after liver transplantation (18). Studies showed that serum AFP values <20 µg/L within 2 months of liver transplantation were indicative of complete tumor removal, and patients with high AFP levels had a higher risk for recurrence (19,20). In our study, we found that the expression of SRC-1 was associated with tumor diameter, TNM grade, and liver cirrhosis, and the expression of Twist1 was correlated with tumor diameter, TNM grade, and liver cirrhosis in HCC patients. In the breast cancer cell line MDA-MB-231, the Twist1 promoter region (–655 to –432 bp) was found to be bound by PEA3 after activation of PEA3 by SRC-1, which ultimately led to an increase in transcriptional activity and expression of Twist1 in breast cancer. Thus, increased Twist1 expression induced by the activation of PEA3 is an important mechanism by which SRC-1 can promote breast cancer metastasis. However, further investigation is still needed to provide deeper insights into these relationships. We demonstrated for the first time that both Twist1 and SRC-1 are independent risk factors for survival in HCC patients. More clinical evidence should be given to further confirm these results.
Conclusions
We analyzed the role of SRC-1 and Twist1 in liver cancer and explored their expression in relation to the prognosis of patients with liver cancer as well as in HCC cell lines. We demonstrated for the first time that both Twist1 and SRC-1 expression are correlated with clinical outcomes and prognosis of HCC patients, and Twist1 and SRC-1, both individually and together are independent risk factors for survival conditions of HCC patients. In addition, silencing SRC-1 inhibited cell proliferation, invasion, migration and EMT in HCC cells, which might occur through inhibition of Twist. This study might give clinical evidence for the value of SRC-1 and Twist1 as prognostic factors and might provide new targets for both the diagnosis and treatment of HCC patients.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.56). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the ethics committee of Guizhou Provincial People’s Hospital [Approval No. EC Review (Animal) 2019-008]. Informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Darvesh AS, Aggarwal BB, Bishayee A. Curcumin and liver cancer: a review. Curr Pharm Biotechnol 2012;13:218-28. [Crossref] [PubMed]
- Osaki Y, Nishikawa H. Treatment for hepatocellular carcinoma in Japan over the last three decades: our experience and published work review. Hepatol Res 2015;45:59-74. [Crossref] [PubMed]
- Kuppusamy P, Nagalingam A, Muniraj N, et al. Concomitant activation of ETS-like transcription factor-1 and death receptor-5 via extracellular signal-regulated kinase in withaferin A-mediated inhibition of hepatocarcinogenesis in mice. Sci Rep 2017;7:17943. [Crossref] [PubMed]
- Ayuso C, Rimola J, Vilana R, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol 2018;101:72-81. [Crossref] [PubMed]
- Dong P, Kaneuchi M, Watari H, et al. MicroRNA-106b modulates epithelial-mesenchymal transition by targeting TWIST1 in invasive endometrial cancer cell lines. Mol Carcinog 2014;53:349-59. [Crossref] [PubMed]
- Li LZ, Zhang CZ, Liu LL, et al. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis 2014;35:469-78. [Crossref] [PubMed]
- Bellovin DI, Tran PT, Adam SJ, et al. Abstract 2975: transgenic mouse model of Twist1-induced metastasis reveals genes highly prognostic for human hepatocellular carcinoma. Cancer Res 2012;72:2975.
- Tan Q, Wang H, Hu Y, et al. Src/STAT3-dependent heme oxygenase-1 induction mediates chemoresistance of breast cancer cells to doxorubicin by promoting autophagy. Cancer Sci 2015;106:1023-32. [Crossref] [PubMed]
- Luef B, Handle F, Santer FR, et al. Abstract 618: implications of inhibition of steroid receptor co-activator-1 in human prostate cancer. Cancer Res 2014;74:618.
- Xu Y, Hu B, Qin L, et al. SRC-1 and Twist1 expression positively correlates with a poor prognosis in human breast cancer. Int J Biol Sci 2014;10:396-403. [Crossref] [PubMed]
- Qin L, Liu Z, Chen H, et al. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res 2009;69:3819-27. [Crossref] [PubMed]
- Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999;17:1474-81. [Crossref] [PubMed]
- Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis 2009;27:80-92. [Crossref] [PubMed]
- Sakamoto M. Early HCC: diagnosis and molecular markers. J Gastroenterol 2009;44:108-11. [Crossref] [PubMed]
- Bourguignon LY, Wong G, Earle C, et al. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J Biol Chem 2010;285:36721-35. [Crossref] [PubMed]
- Lin YJ, Lee MH, Yang HI, et al. Predictability of liver-related seromarkers for the risk of hepatocellular carcinoma in chronic hepatitis B patients. PLoS One 2013;8:e61448. [Crossref] [PubMed]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264-73.e1. [Crossref] [PubMed]
- Nörthen A, Asendorf T, Walson PD, et al. Diagnostic value of alpha-1-fetoprotein (AFP) as a biomarker for hepatocellular carcinoma recurrence after liver transplantation. Clin Biochem 2018;52:20-5. [Crossref] [PubMed]
- Charrière B, Maulat C, Suc B, et al. Contribution of alpha-fetoprotein in liver transplantation for hepatocellular carcinoma. World J Hepatol 2016;8:881-90. [Crossref] [PubMed]
- Yoo T, Lee KW, Yi NJ, et al. Peri-transplant change in AFP level: a useful predictor of hepatocellular carcinoma recurrence following liver transplantation. J Korean Med Sci 2016;31:1049-54. [Crossref] [PubMed]


