Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients
Introduction
Cancers of the head and neck represent an increasingly prevalent disease, with over 600,000 cases occurring each year, accounting for 3.5% of all cancers in the United States (1). Only one-third of patients present with early-stage disease, while the majority will present with advanced disease, often with lymph node metastases (2). Although many patients will be treated with curative intent, a large percentage will expire from their disease due to locoregional recurrence or distant metastases.
The molecular landscape of head and neck cancer (HNC) is complex and currently lacks an armamentarium of effective targeted therapeutics. One exception is cetuximab, a monoclonal antibody specific for epidermal growth factor receptor (EGFR), which is the only targeted therapy approved for use in head and neck squamous cell carcinoma. Cetuximab has been shown to increase survival when added to radiation therapy alone, and has also been shown to increase overall survival when added to traditional platinum-based regimens (3,4). Although the use of targeted therapies is promising, especially in the current medical climate of personalized medicine, more customized treatments are presently unavailable in HNC.
Advanced sequencing technologies have allowed for the understanding that the vast majority of cancers harbor unique somatic mutations. The development of next generation sequencing (NGS) has revolutionized the detection and treatment of malignancies. It is known that circulating tumor DNA (ctDNA) can be present in the blood of advanced cancer patients. Sequencing of blood samples in these patients would allow for a relatively easy way to acquire a “liquid biopsy” of respective tumors and for the identification of unique somatic mutations. This theoretically allows for more personalized treatment with matched therapies through an understanding of the entirety of the patient’s cancer mutanome and avoiding therapeutic issues that arise from tumor heterogeneity or sample limitations. Guardant360 is a commercially available 73-gene panel utilizing digital sequencing of ctDNA isolated from a single, non-invasive peripheral blood draw. The Guardant360 assay has been shown to be both more sensitive and equally as specific as tissue NGS with 85% sensitivity and 99.6% specificity (compared to 80% and 99.7%, respectively) (5). Little is known, however, on the specific utility of such tests in the treatment of recurrent and metastatic (R/M) HNC.
We designed a retrospective study to further characterize the results of blood sample sequencing in R/M HNC patients, to determine its ability to identify actionable mutations, and to further elucidate the utility of blood ctDNA profiling and its role in patient management.
Methods
This study involved a retrospective review of 60 patients with R/M HNC who received care at a single institution between February 2015 and June 2016. Tumor specimen analysis and pertinent clinical data (including tissue and ctDNA genomic testing results) were captured from patient chart review and the study was conducted in accordance with an IRB-approved protocol. The study was approved by UCSD Human Research Protections Program (HRPP 16127). Diagnostic biopsies were performed for tumor specimen analysis as part of routine clinical care at various times throughout the course of treatment, often at the time of diagnosis. Commercially available sequencing platforms were used for tissue sequencing and included both FoundationOne (FoundationOne, Cambridge, Massachusetts, http://www.foundationone.com) and Caris (Caris Life Sciences, Dallas, Texas, https://www.carislifesciences.com). Retrospective review of therapeutic decision making based of ctDNA results was queried and reported.
Blood samples were acquired as part of clinical care at variable times from diagnosis from all patients involved in the study with several patients having multiple blood samples analyzed. Sequencing of ctDNA was performed by Guardant Health Inc. (Guardant360, Redwood City, California, http://www.guardanthealth.com/guardant360/), a Clinical Laboratory Improvement Amendment (CLIA)-certified and College of American Pathologists (CAP)-accredited clinical laboratory (6,7). Two 10 milliliter tubes of blood were acquired from each patient, and 5 to 30 nanograms of ctDNA were isolated from each sample. At the time of this study, the Guardant panel utilized hybrid capture and NGS of critical exons of 70 genes, reporting point mutations for all genes and indels, fusions, and copy number amplifications for select genes (Figure S1) (5). NGS technology enabled variant detection down to 0.02% to 0.04% allelic fraction/2.12 copies with ≤0.3%/2.24–2.76 copies 95% limits of detection with greater-than 98% specificity (6,7). Mutations identified by blood specimen analysis were categorized as actionable or non-actionable. An actionable mutation was defined as having an off-label (non-FDA approved) or clinical trial option identified for an alteration in any of the genes assessed in ctDNA. Mutations uncovered by ctDNA sequencing of blood samples were compared with mutations from tumor specimen DNA sequencing, and the most common mutations found in each platform were identified.
Response Evaluation Criteria in Solid Tumors (RECIST v1.1) measurements were used to calculate change in tumor size and response to treatment in all patients (8). A small subset of patients was identified who received matched targeted therapy (MTT) as a result of ctDNA sequencing and outcomes were compared to those who received standard therapy. Treatment was considered “matched” if the treatment regimen was directly altered as a result of ctDNA sequencing and targeted an alteration in the patient’s molecular profile.
Results
Clinical characteristics
Our cohort consisted of 60 patients; 45 of which were male. The median age of the population was 63 years (Table 1). The majority of patients were classified as non-Hispanic white (n=52, 86.7%) with a smaller subset categorized as Hispanic (n=8, 13.3%). Twenty-one patients (35%) had a significant smoking history of greater than 10 pack years and 19 (31.7%) were never smokers. The most common primary tumor type was oropharyngeal squamous cell carcinoma (SCC) (n=21, 35%), which was also HPV positive 25% (n=15) of the time. Patients with other types of HNC were included in the cohort and included both thyroid (n=4) and salivary gland (n=8) primaries but were not included in specific SCC analyses. At the time of diagnosis, 20% (n=12) of patients initially presented with metastatic disease.
Table 1
| Characteristic | N (%) |
|---|---|
| Sex | |
| Male | 45 (75.0) |
| Female | 15 (25.0) |
| Median age [range] | 63 [44–89] |
| Race | |
| Hispanic | 8 (13.3) |
| Non-Hispanic White | 52 (86.7) |
| Tobacco exposure | |
| Never smoker | 19 (31.7) |
| <10 pack years | 12 (20.0) |
| >10 pack years | 21 (35.0) |
| Unknown | 8 (13.3) |
| HPV status | |
| Positive | 15 (25.0) |
| Negative | 9 (15.0) |
| Unknown | 36 (60.0) |
| Primary site | |
| Oral cavity | 12 (20.0) |
| Oropharynx | 21 (35.0) |
| Nasopharynx | 3 (5.0) |
| Larynx | 6 (10.0) |
| Hypopharynx | 4 (6.7) |
| Salivary gland | 8 (13.3) |
| Thyroid | 4 (6.7) |
| Unknown primary | 2 (3.3) |
| Stage at diagnosis | |
| Tumor stage | |
| T1 | 10 (16.7) |
| T2 | 10 (16.7) |
| T3 | 8 (13.3) |
| T4 | 9 (15.0) |
| Unknown | 23 (38.3) |
| Nodal stage | |
| N0 | 8 (13.3) |
| N1 | 10 (16.7) |
| N2 | 16 (26.7) |
| N3 | 3 (5.0) |
| Unknown | 23 (38.3) |
Mutational analysis
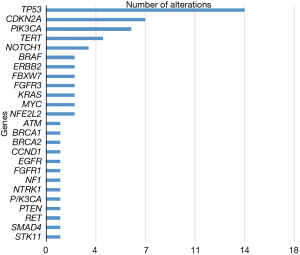
Thirty of the 60 metastatic HNC patients had tissue samples available for comparison. Twenty-nine of these tissue samples were sequenced using FoundationOne, and one was sequenced using Caris. Somatic mutations were identified in 29 samples (96%) and one sample had no detected genetic alterations. The most common mutations identified in tissue were TP53 (48% of patients), CDK2NA (30% of patients), PIK3CA (24% of patients), and FAT1 (16.6% of patients) (Figure 1).
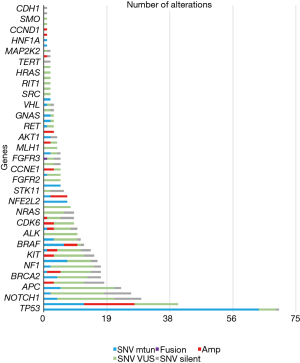
Of the 60 patients in the cohort, 83% (n=50) of patients had at least one alteration identified on ctDNA sequencing. A total of 480 alterations were identified, the majority of which (94%) were point mutations (n=463). The most frequent alterations identified in ctDNA were TP53 (68% of patients), PIK3CA (34% of patients), NOTCH1 (20% of patients), and ARID1A (15% of patients) (Figure 2). Of the 10 patients without detectable mutations on Guardant360, 50% (n=5) had no evidence of disease (NED) and all had survived at the time of follow up.
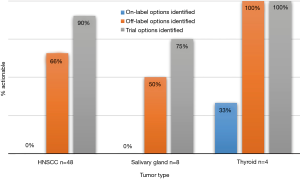
Blood sample ctDNA sequencing identified a new mutation in 73% (n=22) of the patients who also had tissue sequencing. Interestingly, most patients were found to have an actionable alteration, with either an off-label option or clinical trial option identified (Figure 3). In patients with HNC 66% had an off-label option identified and 90% had a trial option identified, while 50% of patients with salivary primaries had off-label options identified and 75% had trial options identified. All patients (n=3) with thyroid primaries had off-label and clinical trial options identified.
Among the 30 patients with tissue sequencing results available, 20 had tissue-detected alterations that were theoretically detectable by the ctDNA assay (i.e., assay panel composition included gene, alteration type, and relevant exon coverage of the tissue-detected mutation). Of the 41 alterations detected in tissue that were covered by the ctDNA assay, 27 (66%) were detected in circulation. Among the 14 not detected, 9 were amplification events, 3 of which were equivocal in tissue, and the remaining 5 included 3 TP53 mutations, one equivocal ERBB2 point mutation, and an FGFR3 point mutation. Focusing on sequence-based mutations, 26 of 31 (84%) point mutations/indels were detected in blood, including detection of BRAF V600E (n=2/2) and activating PIK3CA and KRAS mutations (n=5/5) in 100% of cases. The single fusion event detected in tissue (FGFR3-TACC3) was also detected in ctDNA. Additionally, of the mutations identified solely on blood samples and not in tissue (n=22), 13 had off-label therapeutic target options identified, including ERBB2 (HER2) amplifications (n=2) or activating mutations (n=1), MET amplification (n=2), and PIK3CA activating mutations (n=4) and/or amplification (n=4). Additionally, four somatic homologous recombination gene mutations (2 BRCA1, 1 BRCA2, 1 ATM) were also identified in ctDNA, identifying potential candidates for PARP inhibition therapy.
Response and matched therapy
The outcomes of therapy for patients in this cohort were analyzed utilizing RECIST v1.1. Twenty-six patients (43.3%) had sufficient surveillance imaging for analysis. Of the 34 patients without sufficient imaging, 12 patients (35.3%) were lost to follow up, 12 patients (35.3%) were deceased before treatment could be initiated or scans could be performed, 1 patient (3.0%) was managed surgically, and 9 patients (26.4%) were excluded for other reasons. Of the patients that received subsequent therapy that was not matched, 11 patients had stable disease (52.4%), 9 patients had progressive disease (42.9%), and 1 patient had a complete response (4.8%) on immunotherapy.
A subset of patients were identified who received MTT (13.3%). Best response in these patients included 3 patients with stable disease (37.5%), 3 patients with progressive disease (37.5%), and 2 (25%) were not evaluated due to follow up scans not being completed. Of the patients with stable disease, one with a BRAF mutation was treated with dabrafenib/trametinib, another with a BRAF mutation was treated with vemurafenib, and another was treated with cetuximab as a result of an EGFR mutation. Of those with progressive disease, one patient was treated with everolimus as a result of a PIK3CA mutation, one was treated on a clinical trial with a MET inhibitor, and one was treated with cabozantinib as a result of a RET mutation.
Of the patients who did not receive MTT, the majority of patients (n=9, 45%) expired before treatment could be initiated, 3 (15%) were placed on immunotherapy, 3 had unclear matched therapies, 2 (10%) were placed on active surveillance, 2 (10%) were lost to follow up, and 1 remained stable on their current treatment regimen.
Discussion
This is the first study to investigate the utility of NGS sequencing of blood samples in patients with R/M HNC. The most common mutations identified were TP53 (68% of patients), PIK3CA (34% of patients), NOTCH1 (20% of patients), and ARID1A (15% of patients). Alterations found in tumor NGS were also examined, and the most common alterations identified were TP53 (48% of patients), CDK2NA (30% of patients), PIK3CA (24% of patients), and FAT1 (16.6% of patients), revealing both similarities as well as important differences between the two sequencing modalities. Importantly, 73% of patients with tissue NGS available for comparison had new mutations identified on blood sample sequencing, the majority of which were actionable, and had either an off-label or clinical trial treatment option identified. Additionally, a subset of patients were identified who received MTT, selected as a direct result of their blood sample sequencing. Of these 8 patients, 3 patients had stable disease at follow up, 3 patients had progressive disease, and 2 were not evaluated.
There are undoubtedly many advantages of somatic NGS using blood samples compared to tumor biopsies, as traditional tumor sequencing requires invasive procedures to procure specimens. Depending on the site of recurrence or metastases, prior treatment and patient fitness, serial biopsies can be difficult in this population. In contrast, blood NGS requires only a peripheral blood draw, and has relatively rapid turnaround time and reduced cost. It has been shown that blood ctDNA analysis has high concordance with traditional tumor NGS (9) results and our study supports this. Twenty-seven of 41 tissue detected alterations were also detected in ctDNA (66%), 27/37 (73%) when the tissue equivocal findings were excluded. Twenty-six of 31 (84%) of sequence-based mutations and 5/5 (100%) of the classically actionable mutations (BRAF V600E, FGFR3 fusion) detected in tissue were also identified in ctDNA. Additionally, several studies have demonstrated that non-small cell lung cancer (NSCLC) patients receiving MTT as the result of ctDNA blood sample sequencing have similar response rates and clinical outcomes compared to patients utilizing tumor sequencing, further supporting its utility (10,11). ctDNA has also been used in other studies prognostically, in which genetic changes detected in blood were shown to be predictive of patient outcomes, which supports the potential comprehensive utilization of ctDNA from the time of diagnosis to risk stratification and ultimately treatment (12). Due to ease of specimen collection, relatively rapid turnaround time, reduced cost, and comparable results, the use of liquid biopsy will likely surpass traditional tumor biopsy as the standard practice.
The use of liquid biopsy has also been proposed as a solution to limitations of tumor sampling (13). Multiple studies have illustrated that tumors are, in fact, heterogeneous and that single tumor biopsy specimens only reveal a small subset of alterations present within the entire tumor burden (14,15). Blood sample sequencing may be one way to circumvent this issue, as a more comprehensive sampling method, allowing for the detection of more targetable mutations, as well as more information regarding the tumor’s genetic landscape. It is important to note, however, that not all patients with active malignancies may have mutations detectable in peripheral blood. Fiala et al. suggested that patients would require a tumor volume of at least 10 mm for detection, and therefore patients with early recurrence or a microscopic disease burden may not yet benefit in comparison to those with already widely metastatic disease (12). Additional known reasons for discordance between ctDNA and tissue result include low tumor shed, length of time between tissue and ctDNA sampling, and patient response to therapy (16).
Our study had several limitations. Given the retrospective nature, single center design, and relatively small sample size, there is a possibility for inherent biases within the results. Additionally, tumor samples were collected at variable times from the date of blood sample collection, which may have contributed to differences in alterations detected within respective assays. Furthermore, given the tenuous nature of the health of the R/M HNC population, several patients were excluded from RECIST measurements as well as MTT data as they died before treatment could be initiated or follow up imaging could be performed.
Conclusions
We have described the similarities and differences between tumor NGS and ctDNA sequencing of peripheral blood. There may be a unique role for the use of liquid biopsy in the R/M HNC population. Further studies are needed to elucidate the utility of ctDNA sequencing and its effects on patient outcomes.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.70). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by UCSD Human Research Protections Program (HRPP 16127). Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006;354:567-78. [Crossref] [PubMed]
- Specenier P, Vermorken JB. Cetuximab: its unique place in head and neck cancer treatment. Biologics 2013;7:77-90. [PubMed]
- Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 2015;10:e0140712. [Crossref] [PubMed]
- Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res 2018;24:3539-49. [Crossref] [PubMed]
- Zill OA, Banks KC, Fairclough SR, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018;24:3528-38. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Kim ST, Lee WS, Lanman RB, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget 2015;6:40360-9. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Villaflor V, Won B, Nagy R, et al. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 2016;7:66880-91. [Crossref] [PubMed]
- Fiala C, Kulasingam V, Diamandis E, et al. Circulating Tumor DNA for Early Cancer Detection. JALM 2018;3:300-13. [PubMed]
- Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 2015;7:293ra104. [Crossref] [PubMed]
- Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-92. [Crossref] [PubMed]
- Schwaederlé MC, Patel SP, Husain H, et al. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clin Cancer Res 2017;23:5101-11. [Crossref] [PubMed]