
N6-methyladenosine (m6A) RNA methylation regulators are associated with clinical prognosis in hepatocellular carcinoma
Introduction
In the 1970s, it was discovered that m6A methylation is a post-transcription level regulation (1). It is widely found in different eukaryotes, including yeast, plants, and mammals (2). Due to the low sensitivity of early detection technologies on the m6A site being limited, it was not until 2011 that the first protein associated with demethylase fat mass and obesity (FTO) was clearly identified (3).
There are three known kinds of enzymes that regulate m6A RNA modification: methyltransferases (“writers”), binding proteins (“readers”), and demethylases (“erasers”) (4). It has been widely reported that m6A RNA regulators involve 13 molecules; “writers” including methyltransferase like 3 (METTL3), methyltransferase like 4 (METTL14), RNA-binding motif protein 15 (RBM15), WT1-associated protein (WTAP), zinc finger CCCH domain-containing protein 13 (ZC3H13), KIAA1429, “readers” including heterogeneous nuclear ribonucleoprotein C (HNRNPC), YTH domain-containing 1 (YTHDC1), YTH domain-containing 1 (YTHDC2), YTH N6-methyladenosine RNA-binding protein 1 (YTHDF1), YTH N6-methyl adenosine RNA-binding protein 2 (YTHDF2), and “erasers” including α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5), and FTO (4-10).
m6A methylation, like DNA methylation, can affect tumor progression by regulating the expression levels of tumor suppressor genes or oncogenes (11). m6A methylation is simultaneously associated with cancer stem cells and the response of anti-tumor drugs such as gemcitabine, 5-FU, etc. (12-14). Recent literature has reported that thirteen m6A RNA methylation regulators contribute to malignant progression and have a clinical prognostic impact for gliomas (15). At present, there are few studies on m6A methylation in liver cancer, and the existing studies mainly focus on the biological functions of individual molecules such as KIAA1429 and YTHDF2 (16,17). There are also few integral level analyses of the relationship between m6A RNA methylation regulators and clinical prognosis in hepatocellular carcinoma (HCC).
We therefore systematically analyzed the expression of 13 reported m6A RNA regulators and the clinical characteristic in the Cancer Genome Atlas (TCGA) datasets in this study. We constructed a tumor subgroup model and a risk model to prove that m6A RNA methylation regulators are associated with the clinical prognosis of HCC.
Methods
Datasets and patient samples
RNA-seq transcription data and the corresponding clinical information data were obtained from the TCGA datasets (n=424). RNA-seq transcription data included 50 cases of precancerous tissue and 374 cases of cancer tissue. We extracted the expression data of the thirteen m6A RNA methylation regulators from it. A total of 135 clinical cases were obtained after removing invalid data. Clinical information included age, gender, grade, stage, vascular tumor cell type, Ishak fibrosis score, alpha-fetoprotein (AFP), Eastern Cooperative Oncology Group (ECOG) score, Child-Pugh score, family cancer history, overall survival (OS) time, and survival status. Liver cancer and adjacent tissues from the six HCC patients were collected from the General Hospital of Northern Theater Command. The study protocol was approved by the ethics committee of the General Hospital of Northern Theater Command.
Bioinformatic analysis
We extracted expression data of the 13 m6A RNA methylation regulators from the RNA-sequencing (RNA-seq) transcription data. According to the classification of cancer tissues and adjacent tissues, the Wilcoxon test was used to analyze the differential expression of the m6A RNA methylation regulators. Correlations between m6A RNA methylation regulators were analyzed by the Pearson’s correlation coefficient test.
To clarify the functions of m6A RNA methylation regulators in HCC, we clustered the HCC into different groups using “Consensus Cluster Plus” (50 iterations, resample rate of 80%, and Pearson’s correlation, http://www.bioconductor.org/). Principal component analysis (PCA) analysis was used to evaluate the clustering effects. We combined all of the clinical data to determine the clinical value of the clustering results through clinical relevance analysis and survival analysis.
To clarify the prognosis risk of the genes, we performed a univariate Cox regression analysis of the 13 genes. Based on the result, we constructed a risk model using the LASSO Cox regression algorithm and classified the results into either the high-risk group or the low-risk group. The risk score was calculated using the following formula:
Where Coefi is the coefficient and xi is the expression value of each selected molecule. The receiver operating characteristic (ROC) curve was used to evaluate model accuracy, and multivariate Cox regression was used to analyze the independent prognostic role of the risk model.
In vitro experiment
RIPA buffer containing the protease inhibitor PMSF (Solarbio Science & Technology Company, China) was used to lyse tissues on ice, and BCA kit (Solarbio Science & Technology Company, China) was used for protein quantification. A total of 20 µg proteins were separated by 10% SDS-PAGE and electro-blotted onto nitrocellulose (NC) membrane. After sealing with skimmed milk, the NC membrane was incubated with the first antibody at 4 °C overnight. The membranes were washed and incubated with the second antibody on the shaking table at room temperature for two hours. ECL chemiluminescence kit (Advansta, USA) was used to visualize the protein bands. β-actin was used as a control. The main antibodies used in this study included YTHDF2 (1:1,000) and ZC3H13 (1:1,000) (Abcam, USA).
For mRNA quantifications of YTHDF2 and ZC3H13, cDNA was synthesized by DNase treatment and reverse transcription (TIANGEN Biotech Company, China). Real-time PCR was on TL988 Real-Time PCR Detection System (TIANLONG, China). The primers were listed in Table 1. The mRNA levels of the selected genes were normalized to that of the reference gene β-actin, and the value were calculated by the 2−ΔΔCt method. The results are expressed as the means ± standard error based on three independent experiments.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| YTHDF2 | CTCTTGGAGCAGTACAAAAT | GGGACTGTAGTAACTGGGTA |
| ZC3H13 | AGCAGCAATTATAGAAGGTC | GATTCTTTCCTAACAGGTGA |
| β-actin | ATAGCACAGCCTGGATAGCAACGTAC | CACCTTCTACAATGAGCTGCGTGTG |
Statistical analysis
SPSS 20.0 (SPSS Inc. Chicago, IL, USA) was used for statistical analysis of clinical data. Wilcoxon test was used to compare the differences in each group. A chi-square test was used to analyze the correlation in the different groups. The Kaplan-Meier method was used to compare the OS of the patients in cluster groups or in the high-risk and low-risk groups. Statistical analysis of all RNA-seq transcriptome data was conducted using R v3.4.1 (https://www.r-project.org/). P<0.05 was considered statistically significant.
Results
Expression of m6A RNA methylation regulators in HCC
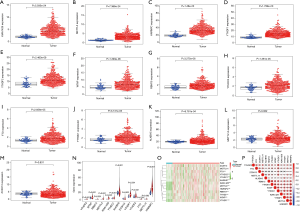
Considering the various biological functions of each m6A RNA methylation regulator on HCC, we analyzed the expression of each molecule in liver cancer and adjacent tissues. The results showed that the expression of mostly m6A RNA methylation regulators was higher in the cancer tissues (P<0.01) (Figure 1A,B,C,D,E,F,G,H,I,J,K), and only METTL14 (P=0.062) and ZC3H13 (P=0.831) were not significantly different (Figure 1L,M). To observe the differential expression of all molecules more intuitively, we plotted the summary maps (Figure 1N,O). The results suggested that m6A methylation may play a significant role in tumorigenesis and development. In addition, we performed a correlation analysis of m6A RNA methylation regulators, and most were positively correlated (Figure 1P).
Consensus clustering of m6A RNA methylation regulators identified two clusters of HCCs with different clinical features
Considering the clustering stability and the number of each group, we divided the patients into two subgroups clustered by k=2, namely, cluster 1 and cluster 2 (Figure 2A,B). The clinical data of the two subgroups clustered are given in Table 2. PCA analysis suggested that the two subgroups clustered had a difference in the expression of m6A RNA methylation regulators (Figure 2C). On that basis, we further compared the clinical features of the two groups. Survival curves showed a significant difference in OS between the two subgroups clustered (P=0.011) (Figure 2D). The 3-year survival rate of cluster 1 was significantly greater than that of cluster 2 (77% vs. 49%). Child-Pugh B, AFP ≥400 µg/L, and low grade were mostly concentrated in cluster 2, indicating a poor clinical outcome (Figure 2E).
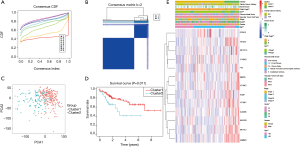
Table 2
| Features | Cluster 1 | Cluster 2 | P value |
|---|---|---|---|
| Total cases | 103 | 32 | – |
| Age | 0.0042 | ||
| <60 years | 39 | 22 | |
| ≥60 years | 64 | 10 | |
| Gender | 0.2209 | ||
| Male | 75 | 19 | |
| Female | 28 | 13 | |
| Grade | 0.0162 | ||
| G1 | 12 | 1 | |
| G2 | 58 | 11 | |
| G3 | 30 | 19 | |
| G4 | 3 | 1 | |
| Stage | 0.7068 | ||
| I | 62 | 19 | |
| II | 24 | 7 | |
| III | 14 | 6 | |
| IV | 3 | 0 | |
| Vascular tumor cell type | 0.3412 | ||
| None | 78 | 20 | |
| Micro | 21 | 10 | |
| Macro | 4 | 2 | |
| Ishak fibrosis score | 0.3518 | ||
| 0 | 33 | 8 | |
| 1,2 | 12 | 6 | |
| 3,4 | 11 | 7 | |
| 5 | 3 | 1 | |
| 6 | 44 | 10 | |
| AFP | 0.0021 | ||
| <400 μg/L | 91 | 20 | |
| ≥400 μg/L | 12 | 12 | |
| Child-Pugh score | 0.0055 | ||
| A | 97 | 24 | |
| B | 6 | 8 | |
| ECOG score | 0.3161 | ||
| 0 | 64 | 18 | |
| 1 | 34 | 10 | |
| 2 | 5 | 4 | |
| Family cancer history | 0.1740 | ||
| No | 65 | 25 | |
| Yes | 38 | 7 | |
Ishak fibrosis score: 0—no fibrosis; 1,2—portal fibrosis; 3,4—fibrous septa; 5—nodular formation and incomplete cirrhosis; 6—established cirrhosis. AFP, alpha-fetoprotein; ECOG, Eastern Cooperative Oncology Group.
Constructing a risk model by using two selected m6A RNA methylation regulators to assess the clinical prognosis of HCC
We next looked for prognostic risk roles of m6A RNA methylation regulators in HCC. Univariate Cox regression analysis suggested that only the expression levels of ZC3H13 and YTHDF2 were related to OS (P<0.05) (Figure 3A). We constructed a LASSO regression model based on the expression of ZC3H13 (Coefi =−0.195) and YTHDF2 (Coefi =0.094) and analyzed the scores among different patients, which were subdivided into high-risk and low-risk groups (Figure 3B). The clinical data of the patients are shown in Table 3. Survival curves showed a significant difference in OS between the two groups (Figure 3C) (P<0.01). The 3-year survival rate and the 5-year survival rate of the high-risk group were less than those of the low-risk group (3-year survival rate: 19% vs. 31%, 5-year survival rate: 12% vs. 17%, respectively). The ROC curve verifies the predictive efficiency of the risk model for survival prediction (Figure 3D). Higher grade was concentrated in the high-risk group, indicating a poor clinical outcome, and the results are in agreement with the subgroup’s analysis (Figure 3E). Univariate and multivariate Cox regression results suggest that the model we constructed can be used as an independent risk factor for predicting the prognosis of HCC (Figure 3F,G).
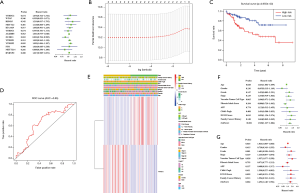
Table 3
| Features | High-risk group | Low-risk group | P value |
|---|---|---|---|
| Total cases | 67 | 68 | – |
| Age | 0.0707 | ||
| <60 years | 36 | 25 | |
| ≥60 years | 31 | 43 | |
| Gender | 0.9547 | ||
| Male | 46 | 48 | |
| Female | 21 | 20 | |
| Grade | 0.0067 | ||
| G1 | 6 | 7 | |
| G2 | 25 | 44 | |
| G3 | 33 | 16 | |
| G4 | 3 | 1 | |
| Stage | 0.2796 | ||
| I | 40 | 41 | |
| II | 16 | 15 | |
| III | 8 | 12 | |
| IV | 3 | 0 | |
| Vascular tumor cell type | 0.1732 | ||
| None | 48 | 50 | |
| Micro | 18 | 13 | |
| Macro | 1 | 5 | |
| Ishak fibrosis score | 0.5562 | ||
| 0 | 18 | 23 | |
| 1,2 | 10 | 8 | |
| 3,4 | 7 | 11 | |
| 5 | 3 | 1 | |
| 6 | 29 | 25 | |
| AFP | 0.4744 | ||
| <400 μg/L | 53 | 58 | |
| ≥400 μg/L | 14 | 10 | |
| Child-Pugh score | 0.3809 | ||
| A | 58 | 63 | |
| B | 9 | 5 | |
| ECOG score | 0.8229 | ||
| 0 | 39 | 43 | |
| 1 | 23 | 21 | |
| 2 | 5 | 4 | |
| Family cancer history | 0.0776 | ||
| No | 50 | 40 | |
| Yes | 17 | 28 | |
Ishak fibrosis score: 0—no fibrosis; 1,2—portal fibrosis; 3,4—fibrous septa; 5—nodular formation and incomplete cirrhosis; 6—established cirrhosis. AFP, alpha-fetoprotein; ECOG, Eastern Cooperative Oncology Group.
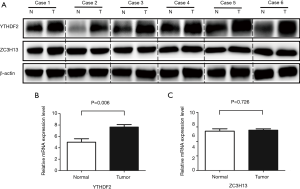
Expression of ZC3H13 and YTHDF2 genes in liver cancer and adjacent tissues
The results were verified by Western blot with in vitro experiment, which can be seen in Figure 4A. Compared with precancerous tissues, the protein level of YTHDF2 in cancer tissues was higher, while the distribution of ZC3H12 protein had no significant difference. We also verified the mRNA level in vitro. With β-actin as a reference, real-time PCR showed that the relative expression of YTHDF2 mRNA was 7.64±0.44, which was higher than precancerous tissues (4.99±0.61) (P=0.006). There was no statistical difference in the expression of ZC3H13 mRNA (5.56±0.18 vs. 5.42±0.33, P>0.05), and those can be seen in Figure 4B,C. The results of the in vitro experiment are consistent with the analysis from TCGA database.
Discussion
Liver cancer is one of the world’s most common malignant tumors, and among all malignant tumors, its mortality ranks third (18), with HCC accounting for 80–90% of all liver cancer (19). The early onset of HCC is not clear, and with a high metastasis level it is common for it to be drug resistant. HCC also has a high rate of recurrence (20). At present, the treatment of HCC is relatively simple. The main means are tyrosine kinase inhibitor (TKI)-targeted therapy such as sorafenib and immunotherapy (21,22). Under a single treatment condition, the prognosis of patients with HCC often differs significantly. A popular research area in the field of oncology is exploring the risk factors affecting the prognosis of HCC (23,24). Conventional risk factors affecting the development of HCC include the Child-Pugh score, Ishak fibrosis score, AFP, family cancer history, etc. (25-27). However, these factors are greatly affected by individual differences; for example, about 31% of HCC patients have an AFP of less than 400 µg/L, and as the age increases, AFP also shows a downward trend (28). The accuracy of using a single factor or multiple factors combined to analyze the prognosis is gloomy, and currently it is impossible to predict the prognosis of liver cancer patients effectively. More sensitive and accurate tumor markers are urgently needed for prognostic stratification and treatment strategy in the development of HCC.
RNA m6A modification refers to a modification in which one hydrogen atom (-H) attached to the sixth nitrogen atom (N6) on the adenine molecule is substituted with a methyl group (-CH4) (29). This modification is widely present in most eukaryotic mRNAs, and the m6A modification is the most abundant endogenous RNA modification (30). m6A modification occurs mostly in polyA mRNA and lncRNA, and is enriched in tissues such as those of the liver and testis (29). m6A modification plays a vital role in oocyte and central nervous system development in early studies (5,31). m6A methylation is involved in tumor progression, drug and radiotherapy resistance, and self-renewal of cancer stem cells in the field of oncology such as colorectal cancer, pancreatic cancer, and glioma (13,32-34).
At present, there are few related studies on analyzing m6A methylation in HCC. Cheng et al. reported that KIAA1429 facilitated migration and invasion of HCC by inhibiting ID2 via up-regulating m6A modification of ID2 mRNA, and Chen et al. reported that METTL3 represses SOCS2 expression in HCC through an m6A-YTHDF2-dependent mechanism (16,17). Most studies have focused on only a single m6A RNA methylation regulator. It is worth mentioning that Zhou et al. confirmed that the combination of METTL3 and YTHDF1 could be regarded as a biological marker that reflects OS in HCC by bioinformatic analysis and clinical verification (35). m6A methylation is enriched in liver tissues, and current research also supports m6A as playing an important role in the occurrence and development of HCC (29). Given these findings and the results of the current study, it is necessary to construct a highly sensitive prognostic prediction model for HCC by combining m6A RNA methylation regulators.
Conclusions
In this study, we analyzed the characteristics of 13 m6A RNA methylation regulators in HCC. We constructed a subtype model and a risk model to prove the correlation between m6A and OS or other clinical features. The subtype model and the risk model we constructed all showed OS differences between the different groups. In addition, both models were associated with clinical features, and the two models complemented each other. Furthermore, Western blot and real-time PCR were used to carry out in vitro experiments, and the results were consistent with those of bioinformatic analysis, which confirmed the validity of our study. m6A RNA methylation regulators are associated with clinical prognosis in HCC. We expect our study can provide a reference for the prognostic stratification and treatment strategy development of HCC.
Acknowledgments
We thank our anonymous reviewers for their valuable comments on this manuscript, which have led to much improvements to the article.
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.84). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the ethics committee of General Hospital of Northern Theater Command [No. k(2016)38]. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hastings MH. m(6)A mRNA methylation: a new circadian pacesetter. Cell 2013;155:740-1. [Crossref] [PubMed]
- Kasowitz SD, Ma J, Anderson SJ, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 2018;14:e1007412. [Crossref] [PubMed]
- Hilário RR, Castro SAB, Ker FTO, et al. Unexpected effects of pigeon-peas (Cajanus cajan) in the restoration of rupestrian fields. Planta Daninha 2011;29:717-23. [Crossref]
- Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. [Crossref] [PubMed]
- Schöller E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA 2018;24:499-512. [Crossref] [PubMed]
- Tang C, Klukovich R, Peng H, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A 2018;115:E325-33. [Crossref] [PubMed]
- Wojtas MN, Pandey RR, Mendel M, et al. Regulation of m6A Transcripts by the 3'→5' RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell 2017;68:374. [Crossref] [PubMed]
- Ding C, Zou Q, Ding J, et al. Increased N6-Methyladenosine causes infertility is associated with FTO expression. J Cell Physiol 2018;233:7055-66. [Crossref] [PubMed]
- Wu R, Yao Y, Jiang Q, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int J Obes (Lond) 2018;42:1378-88. [Crossref] [PubMed]
- Kwok CT, Marshall AD, Rasko JE, et al. Genetic alterations of m 6 A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 2017;10:39. [Crossref] [PubMed]
- Niu Y, Zhao X, Wu YS, et al. N6-methyl-adenosine (m6A) in RNA: An Old Modification with A Novel Epigenetic Function. Genomics, Proteomics & Bioinformatics 2013;11:8-17. [Crossref] [PubMed]
- Cui Q, Shi H, Ye P, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622. [Crossref] [PubMed]
- Taketo K, Konno M, Asai A, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol 2018;52:621-9. [PubMed]
- Zhou S, Bai Z, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog 2018;57:590-7. [Crossref] [PubMed]
- Chai RC, Wu F, Wang QX, et al. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204-25. [Crossref] [PubMed]
- Cheng X, Li M, Rao X, et al. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther 2019;12:3421-8. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Park S, Lim J, Kim JR, et al. Inhibitory effects of resveratrol on hepatitis B virus X protein-induced hepatocellular carcinoma. J Vet Sci 2017;18:419-29. [Crossref] [PubMed]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118-27. [Crossref] [PubMed]
- Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol 2018;19:940-52. [Crossref] [PubMed]
- Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163-73. [Crossref] [PubMed]
- Chen J, Ji T, Zhao J, et al. Sorafenib-Resistant Hepatocellular Carcinoma Stratifiedby Phosphorylated ERK Activates PD-1 Immune Checkpoint. Oncotarget 2016;7:41274-84. [PubMed]
- Miyahara K, Nouso K, Miyake Y, et al. Serum glycan as a prognostic marker in patients with advanced hepatocellular carcinoma treated with sorafenib. Hepatology 2014;59:355-6. [Crossref] [PubMed]
- Hung HH, Chao Y, Chiou YY, et al. A comparison of clinical manifestations and prognoses between patients with hepatocellular carcinoma and child-pugh scores of 5 or 6. Medicine (Baltimore) 2014;93:e348. [Crossref] [PubMed]
- El Raziky M, Attia D, El AW, et al. Hepatic fibrosis and serum alpha-fetoprotein (AFP) as predictors of response to HCV treatment and factors associated with serum AFP normalisation after treatment. Arab J Gastroenterol 2013;14:94-8. [Crossref] [PubMed]
- Everhart JE, Wright EC, Goodman ZD, et al. Prognostic Value of Ishak Fibrosis Stage: Findings from the HALT-C Trial. Hepatology 2010;51:585-94. [Crossref] [PubMed]
- Chen DS, Sung JL. Serum alphafetoprotein in hepatocellular carcinoma. Cancer 1977;40:779-83. [Crossref] [PubMed]
- Fu Y, Dan D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. [Crossref] [PubMed]
- Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187. [Crossref] [PubMed]
- Yang D, Qiao J, Wang G, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res 2018;46:3906-20. [Crossref] [PubMed]
- Nishizawa Y, Konno M, Asai A, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget 2017;9:7476-86. [Crossref] [PubMed]
- Visvanathan A, Patil V, Arora A, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018;37:522-33. [Crossref] [PubMed]
- Dai D, Wang H, Zhu L, et al. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 2018;9:124. [Crossref] [PubMed]
- Zhou Y, Yin Z, Hou B, et al. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag Res 2019;11:3921-31. [Crossref] [PubMed]


