
Circular RNA expression pattern in cisplatin-resistant osteosarcoma cells
Introduction
Osteosarcoma (OS) primarily occurs in the metaphysis of long bones and is the most prevalent primary malignant bone tumor that occurs mostly in children, adolescents, and young adults (1,2). OS commonly presents as pain and swelling in the affected bone. It is estimated that approximately 15% to 20% of patients with OS have tumor metastasis, in which the main target organ is the lungs (2). Due to the benefits of chemotherapy, the long-term survival rate of OS has increased to 70% to 80%, but for patients with metastasis, the 5-year survival rate is still only around 30% (3,4). Moreover, treatment failures are common in OS patients with chemoresistance, worsening patient prognosis (5). Therefore, an in-depth understanding of the disease regulatory mechanism of OS, including chemoresistance, is required to identify relevant diagnostic and therapeutic targets.
Circular RNA (circRNA) is a novel type of endogenous RNA that is derived from precursor mRNA (pre-mRNA) and processed to a circular form by back-splicing. circRNAs are covalently closed structures that lack a canonical 5' cap and 3' polyadenylated tail, making them more resistant to RNase R (6). High-throughput sequencing and microarray-based methods have identified numerous circRNAs in various types of cancers including gastric cancer, ovarian cancer, hepatocellular carcinoma, pancreatic ductal adenocarcinoma, and bladder cancer, as well as OS (7-12). Emerging evidence has indicated that circRNA regulates a wide range of biological processes and modulates tumor behavior, including chemotherapy resistance (11,13,14). Among the circRNAs associated with chemotherapy resistance, circ_0006528 was found to be a regulator of adriamycin resistance in breast cancer (15). Overexpression of two novel circRNAs revealed by RNA sequencing (RNA-seq) significantly enhanced gemcitabine resistance of pancreatic ductal adenocarcinoma cells (16). Two studies revealed the expression pattern of circRNA in OS and chemotherapy resistance OS. Zhu et al. reported the profile of circRNA in chemotherapeutic resistance OS cell lines, and identified hsa_circ_0004674 and hsa_circ_0081001 as chemotherapy resistance-associated circRNAs that could function as biomarkers for OS diagnosis and prognosis (17,18). However, previous studies have not investigated all types of chemotherapy resistance, and circRNA expression in response to cisplatin resistance, which is frequently reported clinically, is not fully understood. The present study used RNA-seq to compare circRNA expression profiles between cisplatin-resistant and cisplatin-sensitive OS cells and aimed to identify the circRNAs expressed in OS tissues. Our findings may provide insight into the mechanisms involved and contribute to targeted therapy screening for cisplatin-resistant OS.
Methods
Cell culture
The U2OS OS cell line was purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (Thermo Fisher Scientific, Life Technologies, MD, USA) and cultured in an incubator with 5% CO2 at 37 °C.
Establishment of cisplatin-resistant cells
U2OS and MG63 cells were cultured at a density of 1×104 cells/well in 96-well plates 12 h prior to cisplatin incubation. After 12 h of culture, cells were then exposed to different concentrations of cisplatin. After 3 days, cell viability was assessed using MTT assay to determine the IC50.
To develop cisplatin-resistant cells, 5×105 U2OS and MG63 cells were incubated with cisplatin at different concentrations, increasing the concentration each week from 0, 0.01, 10, 50, to 100 µM. After 4 weeks of incubation, cisplatin-resistant cell lines of U2OS-R and MG63-R were obtained. Surviving cells were counted and normalized to control cells to validate cisplatin resistance. Resistant cells were cultured continuously in the presence of cisplatin.
Total RNA isolation
U2OS and U2OS-R cells were seeded at a density of 5×105 cells/well in 6-well plates and cultured for 24 h. After 24 h, cells were gently washed with serum-free DMEM. Total RNA was extracted from U2OS and U2OS-R cells using TRIzol reagent (Thermo Fisher Scientific) following a standard protocol. The integrity and size distribution of the total RNA were analyzed using an Agilent 2100 Bioanalyzer pico-RNA chip (Aglient, CA, USA).
Library construction and circRNA-seq
Total RNA samples obtained from U2OS and U2OS-R cells were treated with RQ1 DNase (Promega, USA) to minimize DNA contamination. Next, rRNA was removed from the total RNA samples using the RiboMinus Eukaryote Kit (Qiagen, Hilden, Germen) in accordance with the manufacture’s instruction. Two libraries were created using Next Ultra Directional RNA Library Prep Kit for Illumina (NEB, MA, USA) as per the manufacturer’s instructions. To obtain circRNA reads, libraries were subjected to HiSeq 2000 platform for RNA-seq after quantifying the libraries using an Agilent 2100 Bioanalyzer (Agilent, CA, USA).
Analysis of differentially expressed circRNA
Clean reads obtained from RNA-seq were aligned with the human GRCh37/hg19 reference genome using the Bowtie2 software (bowtie-bio.sourceforge.net/bowtie2). Reads that did not map to the reference genome sequence were selected as the back-splice reads candidates. CIRI software was used to predict circRNA with back-splice algorithm. For differential expression analyses, Reads Per kilobase of exon model per Million mapped reads (RPKM) was used to represent back-splice reads and the reads were normalized to a total read number. Reads that met the criteria of log (fold change) >2 and adjusted P value <0.05 were identified as differentially expressed circRNAs.
To represent an overview of differential expressed circRNAs and to predict their potential function, hierarchical clustering and scatter plots were generated, and gene ontology (GO) analysis was performed using R package 1.0.8 (https://cran.r-project.org/web/packages/pheatmap/). Biological pathway enrichment was analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) database (adjusted P value <0.05; gene count ≥2).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was reverse transcribed using TPrimeScript RT Master Mix (TaKaRa, Dalian, China) for cDNA synthesis. To evaluate the relative expression of circRNAs, cDNA was amplified using a SYBR RT-PCR Master Mix (TOYOBO, Osaka, Japan) kit with divergent primers targeting specific back-splicing site (Table 1). PCR was performed using an ABI 7500 RT-PCR system (Applied Biosystems, CA, USA) and the relative expression of circRNAs was analyzed using the 2–ΔΔCt method. Details of the primers used in the study are shown in Table 2.
Table 1
| circRNA | Chromosome | Gene symbol | Log2 ratio (U2OS-R/U2OS) |
|---|---|---|---|
| hsa_circ_0008336 | chr7:33397467|33427756 | BBS9 | 3.699 |
| hsa_circ_0000375 | chr12:6657591|6657991 | IFFO1 | 3.699 |
| hsa_circ_0085839 | chr8:144547887|144550829 | ZC3H3 | 3.506 |
| hsa_circ_0004188 | chr18:7754371|7774269 | Y_RNA, snoU13, SNORD112, PTPRM | 3.399 |
| hsa_circ_0002419 | chr12:78443773|78452895 | NAV3 | 3.222 |
| hsa_circ_0008749 | chr19:52579297|52592255 | ZNF841 | 3.184 |
| hsa_circ_0002702 | chr9:35546427|35548532 | RUSC2 | 3.158 |
| hsa_circ_0035818 | chr15:64410316|64415745 | SNX1 | 3.158 |
| hsa_circ_0006633 | chr1:59805630|59844509 | FGGY | 3.086 |
| hsa_circ_0003922 | chr2:231307652|231314970 | SP100 | 3.021 |
| hsa_circ_0002809 | chr17:57076742|57094785 | TRIM37 | –2.945 |
| hsa_circ_0046523 | chr17:80828100|80858607 | TBCD | –2.825 |
| hsa_circ_0001493 | chr5:65284463|65310553 | ERBB2IP | –2.693 |
| hsa_circ_0001200 | chr21:46275125|46281186 | PTTG1IP | –2.647 |
| hsa_circ_0087938 | chr9:111868788|111870850 | TMEM245 | –2.623 |
| hsa_circ_0001386 | chr4:1219148|1235307 | CTBP1 | –2.471 |
| hsa_circ_0046580 | chr17:80863812|80869665 | TBCD | –2.417 |
| hsa_circ_0003037 | chr5:14270934|14280545 | TRIO | –2.389 |
| hsa_circ_0005860 | chr7:2252847|2262389 | MAD1L1 | –2.346 |
| hsa_circ_0079149 | chr7:2252847|2259091 | MAD1L1 | –2.301 |
circRNA, circular RNA.
Table 2
| circRNA | Sequence (5'-3') |
|---|---|
| hsa_circ_0008336-F | TTCAACACCTGGACACCTTG |
| hsa_circ_0008336-R | TTTGTGGCTTGCAGTTTTTG |
| hsa_circ_0002809-F | CCCCGGATGAAGATACACAT |
| hsa_circ_0002809-R | GGCTTGGGCGAATTACTCTT |
| hsa_circ_0003302-F | AATGGCAGCGTTGTTACTGA |
| hsa_circ_0003302-R | CCACGGCGATAAGGAAAAT |
| hsa_circ_0004662-F | GCTTGCAAAAAGTAAACCACGA |
| hsa_circ_0004662-R | GCAACTCCCCTTTGGGTTCT |
circRNA, circular RNA.
Statistical analysis
All data were analyzed using SPSS software (version 18.0, SPSS Inc, IL, USA). The difference in cell survival rate was analyzed using Chi-square test and the relative circRNAs expression between normal and resistant cells were analyzed using one-way analysis of variance followed by least significant difference tests. Values of P<0.05 were considered to be significantly different.
Results
Establishment of cisplatin-resistant OS cell lines
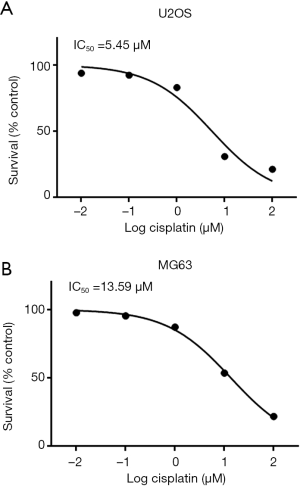
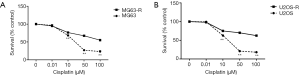
U2OS and MG63 cells were incubated with different concentrations of cisplatin (log cisplatin =–2, –1, 0, 1, and 2 µM) and IC50 values were determined by MTT assay. Cell viability of U2OS and MG63 cells showed a 50% reduction at concentrations of 5.45 and 13.59 µM (Figure 1). Based on the IC50 values of U2OS and MG63 cells, five concentrations (0, 0.01, 10, 50, and 1,000 µM) of cisplatin were selected and the concentration was increased weekly to induced resistant cell lines (U2OS-R and MG63-R). Compared with U2OS and MG63 cells, U2OS-R and MG63-R cells exhibited a higher survival rate after 48 h of incubation with cisplatin. As the concentration increased, the survival rate of U2OS and MG63 cells decreased significantly, whereas only a moderate change was observed in the U2OS-R and MG63-R cells. More than 50% of U2OS-R and MG63-R cells survived after 4 weeks of induction, whereas U2OS and MG63 cells were almost completely dead (Figure 2).
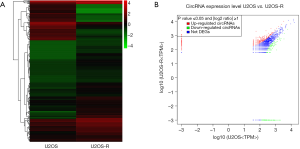
Expression profiles and characteristics of differentially expressed circRNAs in cisplatin-resistant cells
To identify the different characteristics between normal OS cells and cisplatin-resistant cells, circRNA expression profiles of U2OS and U2OS-R cells were compared using RNA-seq. As shown in Figure 3, 343 circRNAs were identified in both cell lines. The expression profiles of circRNAs differed between the U2OS and U2OS-R cells, as demonstrated by hierarchical clustering heat map (Figure 3A). Among the circRNAs, 343 were found to differ in the U2OS-R cells. Among these, 253 were upregulated (red) and 90 were downregulated (green), and could be visualized in the scatter plot (Figure 3B). The top 20 most expressed circRNAs are listed in Table 1.
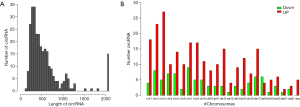
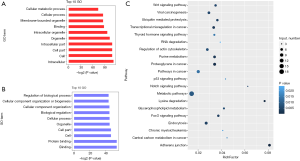
To further explore the characteristics of the circRNAs differentially expressed in the cisplatin-resistant cells, their length and chromosome sites were analyzed. As shown in Figure 3, the length of the differentially expressed circRNAs was enriched in loci around 500 bp (Figure 4A). Upregulated or downregulated circRNAs were observed in all chromosomes, with more circRNAs enriched in chromosomes 1, 2, 3, 7, and 8. There were fewer downregulated circRNAs on the X chromosome (Figure 4B). Differentially expressed circRNAs were subjected to GO and KEGG enrichment pathway analysis. GO analysis revealed that the differentially expressed circRNAs were distributed in biological process, cellular component, and molecular function, and were particularly enriched for cellular metabolic process, cellular process, and membrane-bounded organelle binding for upregulated subsets, while downregulated were enriched for cellular component, organization or biogenesis, and protein binding (Figure 5A,B). Pathway enrichment analysis showed that the differentially expressed circRNAs were enriched in tumor-associated pathways such as metabolic pathways, pathways in cancer, adherens junction, proteoglycans in cancer, and transcriptional misregulation in cancer (Figure 5C).
Identification of four differentially expressed circRNAs in cisplatin-resistant cells
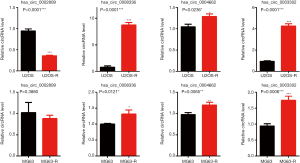
To confirm the data obtained from RNA-seq and to verify the differential expressed circRNAs in the cisplatin-resistant cells, four circRNAs were randomly selected for qPCR confirmatory assay on the U2OS, U2OS-R, MG63, and MG63-R cells. As shown in Figure 6, the relative expression of hsa_circ_0002809, hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 were detected using divergent primers targeting specific to the junction point. Among these four circRNAs, hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 were significantly upregulated, while hsa_circ_0002809 was downregulated in U2OS-R cells compared with U2OS cells. Consistent with these findings, upregulation of hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 was observed in MG63-R cells. A decreased trend in the expression of hsa_circ_0002809 was also observed, although the difference was not statistically significant.
Discussion
The mechanisms of circRNA in cisplatin-resistant OS remain unclear. The findings from the present study provided an overview of circRNA expression in cisplatin-resistant OS cell lines by RNA-seq analysis and support that the expression profile of circRNA in cisplatin-resistant OS cell lines is significantly different from that of cisplatin-sensitive cells. A total of 343 circRNAs were differentially expressed in OS-resistant cell lines. Among these, upregulated circRNAs were mainly concentrated in chromosomes 1, 2, 3, 7, and 8, while the downregulated circRNAs were distributed across all chromosomes, except the X chromosome. Bioinformatics analysis revealed that differentially expressed circRNAs were mainly enriched in metabolic pathways, pathways in cancer-associated pathways such as adherens junction, proteoglycans in cancer, and transcriptional misregulation of cancer pathways.
Previous studies have reported some differentially expressed circRNAs in OS. A series of differentially expressed circRNAs were identified in OS by microarray (19,20) and chemotherapy-associated circRNA expression profiles were shown using high-throughput sequencing (17,18). These findings provide information about circRNA regarding OS progression and therapy study. Among these, some differentially expressed individual circRNAs have been verified by qPCR, including downregulation of circUBAP2 and upregulation of circ0016347, hsa_circ_0001564, circGLI2, hsa_circ_0009910, circNASP, circNT5C2, hsa_circ_008100, and hsa_circ_0004674. Among these, circUBAP2 (21), circ_0001564 (20), and circ_0009910 (22) are considered to have anti-tumor proliferation properties and promote tumor apoptosis. Furthermore, circ0016347 (23), circGLI2 (24), circNASP (25), and circNT5C2 (19) not only have anti-tumor growth effects, but also have inhibitory effects on cell motility, such as tumor migration or invasion. We also confirmed expression of three differentially expressed circRNAs, and showed that hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 were upregulated in cisplatin-resistant OS cells, adding to the number of qPCR-verified circRNAs.
At the RNA level, previous studies on the pathology of OS have mainly focused on miRNAs, lncRNAs, and mRNAs (26). While studies on the disease process of OS from the perspective of circRNA have been reported, these have mainly focused on the tumorigenesis and tumor metastasis. On the other hand, only a few studies have reported circRNAs related to OS chemotherapy resistance (17,18,27). One such study revealed differentially expressed circRNAs between doxorubicin, cisplatin, and methotrexate-resistant patients, reflecting the expression characteristics of resistance-related circRNAs of the three drugs based on chemotherapy. The study identified a total of 80 circRNAs via high-throughput sequencing and verified the high expression of hsa_circ_0004674 in resistant OS tissues (18). Another study revealed the profile of circRNA in chemotherapy-resistant OS cell lines and identified hsa_circ_0081001 as chemotherapy resistance-associated circRNA that could function as a biomarker for OS diagnosis and prognosis (17). A recent study by Zhu et al. revealed a competing endogenous RNA regulatory network between mRNAs and ncRNAs in OS chemoresistance using whole-transcriptome sequencing. They found that hsa_circ_0001258 suppressed doxorubicin resistance of OS cells by upregulating GSTM2 expression via sponging has-miR-744-3p (27). Unlike previous studies, our study focused on cisplatin-resistant OS using a cisplatin-induced cell line. To analyze the expression profile of cisRNA in cisplatin-resistant OS cells, we also verified that hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 were upregulated in cisplatin-resistant OS and downregulated in hsa_circ_0002809. While there are no reports on the functions of these circRNAs, their upregulation in cisplatin-resistant cells suggests that they may be upregulated in response to drug resistance, which may be linked to the development of cisplatin resistance. These mechanisms require further studies.
KEGG analysis of differentially expressed circRNAs showed that upregulated circRNAs in cisplatin-resistant cell lines were mainly concentrated in metabolic pathways, pathways in cancer, adherens junction, proteoglycans in cancer, and transcriptional misregulation in cancer. Compared with a previous study investigating circRNAs associated with chemotherapy resistance (18), we found some differences in glycolysis/gluconeogenesis, ABC transporters, and VEGF signaling pathways. These differences may be due to differences in analysis; therefore, while previous studies investigated three pathways related to drug resistance, we analyzed cisplatin resistance-associated pathways. We found that both metabolic pathways and glycolysis/gluconeogenesis pathways were metabolically related pathways. The regulation of glucose metabolism pathways involved in chemoresistance has been well documented in tumors, including OS (28-31). It is possible that cisplatin resistance may alter the expression of metabolic and glycolysis/gluconeogenesis pathways associated circRNAs, which may contribute to the development of cisplatin resistance. Further circRNA functional studies may provide more evidence.
Taken together, we demonstrated the expression profile of circRNA in cisplatin-resistant OS cells and verified that hsa_circ_0008336, hsa_circ_0004664, and hsa_circ_0003302 were upregulated in OS cell lines, suggesting that these three circRNAs may be resistant to OS. Functional analysis of these circRNAs and their potential use as diagnostic markers will deepen our understanding of the mechanisms by which circRNAs regulate cisplatin resistance in OS cells.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.80). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002;20:776-90. [Crossref] [PubMed]
- Isakoff MS, Bielack SS, Meltzer P, et al. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol 2015;33:3029-35. [Crossref] [PubMed]
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009;152:3-13. [Crossref] [PubMed]
- Stiller CA. International patterns of cancer incidence in adolescents. Cancer Treat Rev 2007;33:631-45. [Crossref] [PubMed]
- Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther 2006;6:1075-85. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132-6. [Crossref] [PubMed]
- Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 2015;5:8057. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 2018;418:41-50. [Crossref] [PubMed]
- Qu S, Song W, Yang X, et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data 2015;5:385-7. [Crossref] [PubMed]
- Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 2017;24:357-70. [Crossref] [PubMed]
- Liu Y, Dong Y, Zhao L, et al. Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol 2018;53:1752-62. [PubMed]
- Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017;9:1175-88. [Crossref] [PubMed]
- Shao F, Huang M, Meng F, et al. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol 2018;9:584. [Crossref] [PubMed]
- Kun-Peng Z, Chun-Lin Z, Jian-Ping H, et al. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci 2018;14:1513-20. [Crossref] [PubMed]
- Kun-Pen Z, Xiao-Long M, Lei Z, et al. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 2018;10:1327-46. [Crossref] [PubMed]
- Liu X, Zhong Y, Li J, et al. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget 2017;8:114829-38. [PubMed]
- Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun 2018;495:2369-75. [Crossref] [PubMed]
- Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 2017;8:61687-97. [PubMed]
- Deng N, Li L, Gao J, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun 2018;495:189-96. [Crossref] [PubMed]
- Jin H, Jin X, Zhang H, et al. Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget 2017;8:25571-81. [PubMed]
- Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol 2017;39:1010428317709991. [Crossref] [PubMed]
- Huang L, Chen M, Pan J, et al. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun 2018;500:511-7. [Crossref] [PubMed]
- Xie L, Yao Z, Zhang Y, et al. Deep RNA sequencing reveals the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma tumorigenesis and pulmonary metastasis. Cell Death Dis 2018;9:772. [Crossref] [PubMed]
- Zhu KP, Zhang CL, Ma XL, et al. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA Networks in osteosarcoma chemo-resistance. Mol Ther 2019;27:518-30. [Crossref] [PubMed]
- Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys Acta Rev Cancer 2017;1868:1-6. [Crossref] [PubMed]
- Veldman RJ, Klappe K, Hinrichs J, et al. Altered sphingolipid metabolism in multidrug-resistant ovarian cancer cells is due to uncoupling of glycolipid biosynthesis in the Golgi apparatus. FASEB J 2002;16:1111-3. [Crossref] [PubMed]
- Xu Y, Gao W, Zhang Y, et al. ABT737 reverses cisplatin resistance by targeting glucose metabolism of human ovarian cancer cells. Int J Oncol 2018;53:1055-68. [PubMed]
- Sullivan EJ, Kurtoglu M, Brenneman R, et al. Targeting cisplatin-resistant human tumor cells with metabolic inhibitors. Cancer Chemother Pharmacol 2014;73:417-27. [Crossref] [PubMed]


