
Expression of MTERF3 gene in breast carcinoma and the relationship with clinicopathological characteristics
Introduction
In recent decades, malignant tumors have become the second leading cause of death worldwide, seriously endangering people's physical and mental health (1). Among all cancerous diseases, breast carcinoma is the most commonly malignant tumor in the world and is also the main cause of death among women under 45 years old in China (2). Compared with other cancers, breast carcinoma patients tend to be younger. Due to the improvement of treatment and the diversification of treatment methods, the 5-year overall survival (OS) rate of breast cancer has improved in the past three years. The corresponding survival rates of breast cancer in 5, 10 and 15 years were 89%, 83% and 78%, respectively. However, there are still some patients with poor prognosis, and the incidence is increasing annually (3). With the in-depth study of breast cancer, it has been demonstrated that the growth and invasion of breast cancer are closely related to a variety of genes (4). Therefore, it is necessary to illustrate the mechanisms for breast cancer tumorigenesis and progression, and it is also urgent to explore novel prognostic markers for developing new effective therapeutic targets for breast cancer.
Mitochondria, an important organelle in eukaryotic cells, play a key role in the regulation of cell energy metabolism, biosynthesis and cell death (including apoptosis and programmed necrosis) (5-7). Because mitochondria are essential for maintaining the homeostasis of life, the dysfunction of mitochondria is related to many human diseases, such as neurodegenerative diseases, cancer, heart disease and diabetes (8-10). Mitochondrial transcription termination factors (mTERFs) are a superfamily of proteins encoded by nuclear genes, that are transported form the nucleus to the cytoplasm, and localize to mitochondria, which have specific mTERF domains (30 amino acid-repeat leucine zipper structures) (11). mTERFs participate in the regulation of mitochondrial gene replication, transcription and translation by binding with mitochondria (12).
The human mitochondrial transcription termination factor 3 (MTERF3) protein, the most highly conserved member of the human MTERF protein family, consists of 417 amino acid residues and 5 conserved mTERF motifs (13). Previous studies have shown that mammalian MTERF3 is a negative regulator of mitochondrial DNA (mtDNA) transcription, it binds the promoter region of mtDNA, which results in the inhibition of double-stranded mtDNA transcription initiation, thereby slowing down cellular energy production (14). Furthermore, MTERF3 is necessary for mammalian embryonic development, knockout of MTERF3 causes a delay in embryonic development and embryonic lethality (14). The inactivation of MTERF3 in the myocardium and skeletal muscle tissue may lead to abnormal mtDNA transcription, resulting in severe respiratory chain defects and decreased oxidative phosphorylation (15). MTERF3 gene amplification and overexpression are common in various types of solid tumors (16). In addition, abnormal protein expression of MTERF3 may be involved in the occurrence, development, and metastasis of malignant tumors (16-18). However, the role of MTERF3 gene in breast cancer has remained elusive. Therefore, an in-depth bioinformatics analysis associated with MTERF3 and breast cancer patients is required. This study analyzed the clinicopathological and survival data associated with MTERF3 expression in breast cancer patients, based on the public cancer database and clinical breast samples.
Methods
Cell lines and culture conditions
The MCF7 (Luminal A), BT-474 (Luminal B), SKBR3 (HER2 overexpression), MDA-MB-468 (Basal like), and MCF10A cell lines were obtained from American Type Culture Collection (ATCC, 2012) and grown in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco BRL) as recommended. All cells were authenticated via STR profiling. Mycoplasma was often checked using the TransDetect Luciferase Mycoplasma Detection Kit purchased from Beijing TransGen Biotech.
Patients and tissue specimens
Breast cancer specimens (n=58) and noncancerous breast tissues specimens (n=58) were collected at the Thyroid and Breast Surgery of the First People’s Hospital of Dali Autonomous Prefecture from June 2013 to December 2015. All of the specimens were confirmed by the Pathology Department and were placed in a −80 °C ultralow temperature freezer within 30 min after surgical resection for cryopreservation. The samples collected included 15 cases of ductal carcinoma in situ, 20 cases of invasive ductal carcinoma, 8 cases of mucinous carcinoma, 10 cases of invasive lobular carcinoma, and 5 cases of mixed carcinoma (intraductal and lobular carcinoma). Of the 58 patients, there were 57 females and 1 male, aged 29–68 years, with a median age of 50.16 years. All the patients were diagnosed for the first time and did not have a treatment history of chemotherapy or radiotherapy before surgery. The study was approved by the Medical Ethics Committee of Dali University (No. 2014-12), and all patients signed an informed consent form.
Immunohistochemistry (IHC) analysis
IHC was performed using a ready-to-use SP kit (Fujian Maxin Biotech Co., Ltd, Fujian, China). The breast cancer and noncancerous breast tissues samples were immediately fixed in 10% neutral buffered formalin and subsequently embedded in paraffin. The paraffin-embedded tissues were sliced continuously at 4 µm. Then the tissues sliced were baked in an oven at 60 °C for 15 min. The slices were dewaxed with xylene and hydrated with gradient ethanol. The endogenous peroxidase was blocked with, 3% hydrogen peroxide for 10 min. Following fully rinsing with PBS, the slices were immersed in citric acid buffer at pH 6.0 and were microwaved for antigen thermal retrieval for 10 min, followed by cooling at room temperature for 20 min. After a second wash with PBS for 10 min, the rabbit anti-human MTERF3 polyclonal antibody (Cat# ab230232, Abcam, Cambridge, MA, USA) that had been diluted to 1:200 was added to the slides and incubated in a refrigerator at 4 °C overnight. Then, the slides were washed with PBS three times (three min each); the goat anti-rabbit IgG (Beyotime Biotechnology Co., Ltd., Shanghai, China) was applied and incubated at room temperature for 30 min. Then, the slides were washed with PBS three times (three min each). The DAB (Fujian Maxin Biotech Co., Ltd, Fujian, China) chromogenic agent was added and incubated for 10 min, with the development monitored under a microscope, and the reaction was stopped by rinsing with distilled water. After using hematoxylin for counterstaining and fully rinsing with distilled water, 1% ethanol-hydrochloric acid was used for differentiation, followed by thoroughly washing with distilled water. Cell staining was observed under a microscope after routine dehydration, xylene treatment, and mounting with neutral resin.
Evaluation of immunostaining results
The intensity of the immunostaining was scored separately by three independent experienced pathologists, who were blinded to the clinicopathological data and clinical outcomes of the patients. The scores of the three pathologists were compared and any discrepant scores were resolved through the re-examination of the staining by the pathologists to achieve a consensus score. The immunolabeling of cancer cells was evaluated. At high microscopy magnification (400×), five representative fields of view were selected, and the proportion of positively staining cells was counted in each visual field. According to the antibody specification sheet, the expression of MTERF3 protein was defined as brownish-yellow staining in the cytoplasm. A value for MTERF3 staining intensity were assigned as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The values of percentage of positive tumor cells were scored as follows: 1 (0–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%). The immunoreactive score (IRS) of each section was calculated by the product of the staining intensity and percentage of tumor cells. According to the IRS, staining patterns were divided into three classes: weak (IRS: 0–3), moderate (IRS: 4–7), and strong (IRS: 8–12) (19).
Determination of MTERF3 mRNA expression by semiquantitative RT-PCR
Primer Premier 5.0 software was used to design PCR primers according to sequences provided by GenBank, and the primer homology was compared using the NCBI/Primer-BLAST website (20). A 100 mg frozen breast cancer and noncancerous breast tissues samples was used for the isolation of total RNA, with TRIzol solution. Total RNA was extracted with a one-step method, according to the instructions of the TRIzol kit (Thermo Fisher Scientific, Inc., MA, USA), and then cDNA was synthesized according to the manufacturer’s instructions (GeneCopeia, Inc., USA). cDNA was used as a template for PCR, the PCR system were cDNA 1 µL, Forward 1 µL, Reverse 1 µL, 2× Taq PCR Master Mix (Beyotime Biotechnology Co., Ltd., Shanghai, China) 12.5 µL, ddH2O 9.5 µL, total 25 µL, and the PCR conditions were as follows: pre-denaturation at 95 °C for 5 min; 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, for a total of 30 cycles, followed by extension at 72 °C for 5 min. The primer sequences of the target gene MTERF3 were MTERF3-F: 5'-ATATCCTCTGACAATTGCT-3', and MTERF3-R: 5'-GAATGATCCACATAGTCTCG-3'; the expected band was 320 bp. The primer sequences of the reference gene GAPDH were GAPDH-F: 5'-GTCAGTGGTGGACCTGACCT-3', GAPDH-R: 5'-TGCTGTAGCCAAATTCGTTG-3', with an expected amplified fragment length of 245 bp. After agarose gel electrophoresis, the PCR products were imaged with a ChemiDoc XRS+ gel imaging system (Bio-Rad, Inc., USA) and analyzed with the Image LabTM 5.2.1 software (Bio-Rad, Inc., USA). mRNA expression of MTERF3 is expressed as the ratio of the accumulated optical density between the target band and the internal reference band. The experiment was repeated three times.
Protein isolation and Western blotting
Protein samples were harvested from 58 breast cancer tissue and 58 noncancerous breast tissue lysates, and the protein concentration was determined by using a BCA Protein Assay Kit (Thermo Fisher Scientific, Inc., MA, USA). The proteins were separated by 12% SDS-PAGE and transferred onto poluvinylidene difluoride membrane (Immobilon, pore size 0.45 µm, Millipore, Bedford, USA). The membrane was blocked with 5% fat-free milk in PBS for 2 h at room temperature and then incubated overnight at 4 °C with rabbit anti-human MTERF3 polyclonal antibody (Cat# ab230232, Abcam, Cambridge, MA, USA). After washing with PBS, the membrane was probed with the corresponding secondary antibody coupled to horseradish peroxidase (Cat# ab6721, Abcam, Cambridge, MA, USA) for 2 h at room temperature. Finally, the protein signals were detected with TanoTM Hig-sig ECL (Beyotime Biotechnology Co., Ltd., Shanghai, China) Western Blotting Substrate and analyzed with the Image LabTM 5.2.1 software (Bio-Rad, Inc., USA). The protein expression of MTERF3 is expressed as the ratio of the accumulated optical density between the target band and the internal reference band. The experiment was repeated three times.
Data collection of breast cancer in TCGA database
The mRNA expression level of MTERF3 (Seq V2) data were retrieved from the Cancer Genome Atlas (TCGA) database (http://tcga-data.nih.gov/tcga/) by R3.6.1 (Cgdsr package) software. The level 3 TCGA data (brca_tcga_rna_seqv2_mrna) were obtained at the TCGA database. Meanwhile, the clinical data, cumulative OS and disease free survival (DFS) data corresponding to MTERF3 mRNA expression level were downloaded for the correlation analysis of clinicopathological parameters and prognosis analysis.
Screening of a breast cancer dataset in TCGA and the correlation analysis of clinical pathological parameters
The data of a total 1,110 breast cancer tissue samples containing MTERF3 mRNA expression were downloaded by using the R3.6.1 software, and 1,110 breast cancer cases had corresponding clinicopathological data and prognostic data. For the analysis of survival and follow-up, the date of surgery was used as the beginning of the follow-up period. The analysis endpoint for the cohort of patients was the time to death or the date of last visit (if death did not occur). The screening and analysis of the downloaded clinical case information was used to exclude cases with incomplete pathological parameters and cases without a prognostic follow-up. After case selection, 705 cases with complete clinical parameters and survival data were incorporated. According to second-generation mRNA-sequencing data, the expression of the MTERF3 mRNA in breast cancer ranged from 105.6388–2392.8571, with a median of 414.115. If the expression of the MTERF3 mRNA in breast cancer sample was greater than the median, that sample was defined as having high expression of MTERF3 (728.30±319.51); otherwise, the sample was defined as having low expression of MTERF3 (299.34±62.38). Accordingly, the MTERF3 mRNA was highly expressed in 352 cases and lowly expressed in 353 cases. The clinical patient data from TCGA database, such as age, sex, tumor status, nodal status, metastasis status, AJCC stage, ER status, PR status, HER2 status, breast cancer molecular typing, cancer type, histological diagnosis, primary site, MTERF3 expression, survival time and outcome, were quantitatively assigned after the establishment of clinical database for the patients.
Molecular typing of breast cancer
According to the molecular typing criteria of breast cancer (21), the pathology of breast cancer sample was divided used to assign the following four molecular types. (I) Luminal A: estrogen receptor (ER)- and/or progesterone receptor (PR)-positive (+), human epidermal growth factor receptor 2 (HER2)-negative (−) and Ki-67 (marker of proliferation Ki-67) <14%; (II) Luminal B: ER and/or PR (+), HER2 (+) and Ki-67 >14%; (III) HER2 overexpression: ER (−), PR (−) and HER2 (+); and (IV) Basal like: ER (−), PR (−), HER2 (−), CK5/6 (+) and/or epidermal growth factor receptor (EGFR) (+).
Correlation between MTERF3 and other mitochondrial regulatory genes in breast cancer
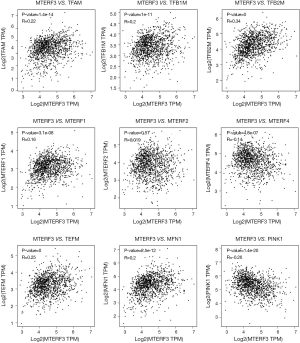
The Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) was used to analyze the correlation between MTERF3 and other gene expression levels related to the present project on the TCGA database and the Genotype-Tissue Expression (GTEx) projects. This study analyzed the breast cancer dataset downloaded from TCGA. The selection of mitochondrial regulatory genes included the following: mitochondrial transcription factor A (TFAM); mitochondrial transcription factor B1 (TFB1M); mitochondrial transcription factor B2 (TFB2M); mitochondrial transcription termination factor 1 (MTERF1); mitochondrial transcription termination factor 2 (MTERF2); mitochondrial transcription termination factor 4 (MTERF4); mitochondrial transcription elongation factor (TEFM); mitofusin 1 (MFN1); and PTEN-induced putative kinase 1 (PINK1).
Statistical analysis
The expression level of MTERF3 in breast cancer tissues was compared with a Mann-Whitney U nonparametric test. Measurement data are expressed as the mean± standard deviation (x ± s), and categorical data are presented as the frequency (percentage). The expression level of MTERF3 in breast cancer tissues was evaluated with Shapiro-Wilk test. The results showed that the expression level of MTERF3 did not have a normal distribution. The correlation between MTERF3 expression and clinicopathological factors was evaluated with a χ2 test and Fisher’s exact probability test. Kaplan-Meier analysis was used to evaluate the effect of MTERF3 on the survival of breast cancer patients, while the log-rank test was used to compare the survival rates. Further, Cox proportional hazards model was used to analyze the possible risk factors affecting the prognosis of patients with breast cancer. Pearson’s correlation analysis was used to analyze the relationship between the MTERF3 expression level with other genes in breast cancer patients. R3.6.1 (EpiDisplay package) and GraphPad Prism 5.01 software were used for statistical analysis and mapping. P<0.05 and P<0.01 indicated significant differences and highly significant differences, respectively.
Results
MTERF3 is upregulated in breast cancer cells
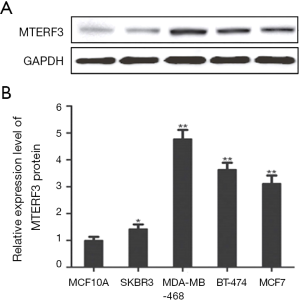
Western blotting results revealed that the protein expression of MTERF3 was highest in MDA-MB-468 cell (Basal like) and lowest in SKBR3 cell (HER2 overexpression), and the protein expression of MTERF3 were significantly upregulated in diverse breast cancer cells including MDA-MB-468 cell (Basal like), SKBR3 cell (HER2 overexpression), MCF7 cell (Luminal A), and BT-474 cell (Luminal B), compared with MCF10A (Figure 1).
MTERF3 is upregulated in breast cancer tissues
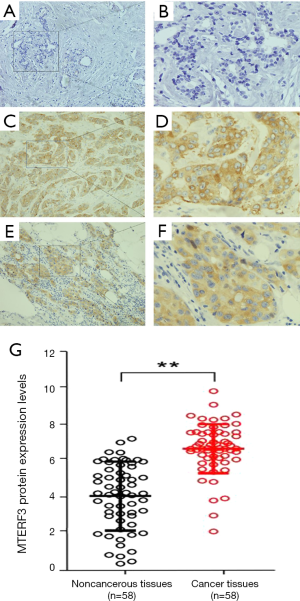
Immunohistochemical staining results showed that the positively stained MTERF3 proteins are fine brown-yellow granules and localized in the cytoplasm of breast cancer. Strong MTERF3 staining was detected in breast cancer tissues, whereas absent or low expression was showed in most noncancerous breast tissues. Compared with noncancerous breast tissues to the tumor of the same patient, the breast cancer tissues exhibited highly increased expression levels of MTERF3 protein (Figure 2). Of the 58 breast cancer specimens, 53 were positive for MTERF3, and the positivity rate was 91.38% (53/58). The positivity rate of MTERF3 was 32.76% (19/58) in noncancerous breast tissues. Therefore, these data showed there was a significant difference between the two groups (P<0.01).
mRNA and protein expression of MTERF3 are significantly increased in breast cancer tissues
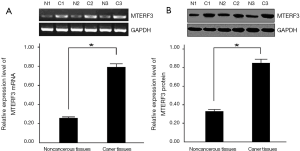
Semiquantitative RT-PCR showed that the mRNA expression level of MTERF3 in 58 breast cancer tissues was significantly higher than noncancerous breast tissues to the tumor from the same samples (P<0.05) (Figure 3A ). Western blot results also demonstrated that the protein expression level of MTERF3 in breast cancer tissues was significantly increased compared with noncancerous breast tissues (P<0.05) (Figure 3B).
Elevated mRNA expression of MTERF3 is correlated with the clinicopathological features of breast cancer patients
In the TCGA breast cancer dataset, the transcription level of MTERF3 was low in 353 cases and high in 352 cases. The mRNA expression level of MTERF3 was significantly correlated with the ER status (P<0.001), PR status (P<0.001), breast cancer molecular typing (P<0.001), cancer type (P<0.001), histological diagnosis (P<0.001), and primary site (P=0.031). The mRNA expression level of MTERF3 was not correlated with age, sex, tumor status, nodal status, metastasis status, AJCC stage, HER2 status, and history of other malignancies (P>0.05) (Table 1).
Table 1
| Clinical indicators | Total | MTERF3 expression | P | χ2 | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age | 0.845 | 0.04 | |||
| <60 | 400 | 199 | 201 | ||
| ≥60 | 305 | 154 | 151 | ||
| Sex | 0.474 | 0.51 | |||
| Female | 697 | 350 | 347 | ||
| Male | 8 | 3 | 5 | ||
| Tumor status | 0.155 | 3.72 | |||
| T1 | 172 | 92 | 80 | ||
| T2 | 422 | 199 | 223 | ||
| T3 + T4 | 111 | 62 | 49 | ||
| Nodal status | 0.349 | 4.45 | |||
| N0 | 343 | 164 | 179 | ||
| N1 | 226 | 125 | 101 | ||
| N2 | 84 | 42 | 42 | ||
| N3 | 47 | 20 | 27 | ||
| NX | 5 | 2 | 3 | ||
| Metastasis status | 0.899 | 0.21 | |||
| M0 | 607 | 303 | 304 | ||
| M1 | 9 | 4 | 5 | ||
| MX | 89 | 46 | 43 | ||
| AJCC stage | 0.607 | 1.00 | |||
| Stage I | 116 | 63 | 53 | ||
| Stage II | 414 | 204 | 210 | ||
| Stage III + IV | 175 | 86 | 89 | ||
| ER | <0.001 | 69.38 | |||
| Negative | 158 | 33 | 125 | ||
| Positive | 547 | 320 | 227 | ||
| PR | <0.001 | 39.25 | |||
| Negative | 234 | 78 | 156 | ||
| Positive | 471 | 275 | 196 | ||
| HER2 | 0.172 | 1.87 | |||
| Negative | 544 | 280 | 264 | ||
| Positive | 161 | 73 | 88 | ||
| Molecular type | <0.001 | 76.37 | |||
| Luminal A | 433 | 263 | 170 | ||
| Luminal B | 125 | 60 | 65 | ||
| HER2 overexpression | 36 | 13 | 23 | ||
| Basal-like | 111 | 17 | 94 | ||
| Cancer type | <0.001 | 44.37 | |||
| Breast invasive ductal carcinoma | 520 | 224 | 296 | ||
| Breast invasive lobular carcinoma | 130 | 98 | 32 | ||
| Other cancer type | 55 | 31 | 24 | ||
| Histological diagnosis | <0.001 | 46.97 | |||
| Infiltrating ductal carcinoma | 507 | 214 | 293 | ||
| Infiltrating lobular carcinoma | 128 | 95 | 33 | ||
| Other histological diagnosis | 70 | 44 | 26 | ||
| History other malignancy | 0.416 | 0.66 | |||
| No | 664 | 335 | 329 | ||
| Yes | 41 | 18 | 23 | ||
| Primary site | 0.031 | 4.64 | |||
| Left | 376 | 174 | 202 | ||
| Right | 329 | 179 | 150 | ||
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Elevated mRNA expression of MTERF3 is not associated with prognosis of breast cancer patients
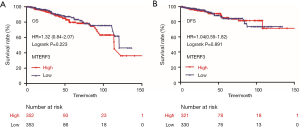
The OS rate and DFS rate were analyzed with the Kaplan-Meier method and the follow-up data of patients with high and low mRNA expression of MTERF3. Kaplan-Meier analysis showed that MTERF3 expression was not associated with the OS of breast cancer patients (HR =1.32, 95% CI: 0.84–2.07, log-rank P=0.223) (Figure 4A). In addition, the expression level of MTERF3 mRNA was not related to the DFS of breast cancer patients (HR =1.04, 95% CI: 0.59–1.82, log-rank P=0.891) (Figure 4B). There was no significant difference between breast cancer patients with high or low MTERF3 expression in regard to the OS time (36.42±31.17 vs. 34.24±29.99 months) and disease-free progression survival time (34.00±29.77 vs. 32.59±28.68 months).
Prognostic value of clinicopathological parameters in breast cancer patients
Univariate and multivariate analyses were performed to identify factors influencing the survival of breast cancer patients. In the univariate analysis, factors such as age, tumor status, nodal status, metastasis status, AJCC stage, cancer type and the history if other malignancies influenced the prognosis of patients (P<0.05), whereas the ER status, PR status, HER2 status, histological diagnosis, primary site and the expression of MTERF3 had no influence on the prognosis (P>0.05) (Table 2).
Table 2
| Factors | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age | |||||||
| ≥60 vs. <60 | 2.65 | 1.68–4.16 | <0.001 | 3.22 | 1.91–5.42 | <0.001 | |
| Tumor status (T2 or T3 + T4 vs. T1) | |||||||
| T2 | 1.03 | 0.59–1.80 | 0.908 | 0.83 | 0.34–2.05 | 0.691 | |
| T3 + T4 | 2.33 | 1.25–4.35 | 0.008 | 1.47 | 0.49–4.44 | 0.491 | |
| Nodal status (N1 or N2 or N3 or NX vs. N0) | |||||||
| N1 | 1.40 | 0.82–2.39 | 0.217 | 1.12 | 0.51–2.45 | 0.786 | |
| N2 | 2.03 | 1.03–4.01 | 0.042 | 1.12 | 0.31–4.14 | 0.860 | |
| N3 | 5.28 | 2.33–11.97 | <0.001 | 2.01 | 0.55–7.41 | 0.292 | |
| NX | 3.97 | 1.11–13.00 | 0.034 | 2.00 | 0.54–7.36 | 0.297 | |
| Metastasis status (M1 or MX vs. M0+cM0) | |||||||
| M1 | 7.89 | 3.39–18.35 | <0.001 | 2.33 | 0.76–7.14 | 0.140 | |
| MX | 0.48 | 0.15–1.55 | 0.221 | 0.26 | 0.07–0.88 | 0.030 | |
| AJCC stage (stage II or stage III + IV vs. stage I) | |||||||
| Stage II | 1.05 | 0.53–2.08 | 0.884 | 1.36 | 0.42–4.44 | 0.612 | |
| Stage III + IV | 3.08 | 1.55–6.13 | 0.001 | 2.93 | 0.48–17.88 | 0.243 | |
| ER | |||||||
| Positive vs. negative | 0.61 | 0.38–1.03 | 0.053 | 0.55 | 0.25–1.23 | 0.147 | |
| PR | |||||||
| Positive vs. negative | 0.75 | 0.47–1.18 | 0.210 | 0.76 | 0.36–1.61 | 0.479 | |
| HER2 overexpression | |||||||
| Positive vs. negative | 1.57 | 0.96–2.59 | 0.074 | 1.18 | 0.67–2.07 | 0.570 | |
| Cancer type (breast invasive lobular carcinoma or other cancer type vs. breast invasive ductal carcinoma) | |||||||
| Breast invasive lobular carcinoma | 0.93 | 0.50–1.74 | 0.826 | 0.12 | 0.02–0.98 | 0.048 | |
| Other cancer type | 2.15 | 1.12–4.13 | 0.022 | 2.54 | 0.75–8.63 | 0.135 | |
| Histological diagnosis (infiltrating lobular carcinoma or other histology types vs. infiltrating ductal carcinoma) | |||||||
| Infiltrating lobular carcinoma | 0.98 | 0.53–1.80 | 0.947 | 7.28 | 0.99–53.28 | 0.051 | |
| Other histology types | 1.46 | 0.76–2.81 | 0.259 | 0.67 | 0.21–2.17 | 0.505 | |
| History other malignancy | |||||||
| Yes vs. no | 2.20 | 1.00–4.82 | 0.049 | 1.93 | 0.78–4.79 | 0.154 | |
| Primary site | |||||||
| Right vs. left | 1.08 | 0.69–1.69 | 0.732 | 1.02 | 0.63–1.65 | 0.944 | |
| MTERF3 | |||||||
| High vs. low | 1.26 | 0.80–1.98 | 0.319 | 1.03 | 0.61–1.71 | 0.923 | |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
A Cox regression model was used to analyze these factors. The results showed that the age, metastasis status and cancer type of breast cancer patients were independent factors leading to the poor prognosis of breast cancer patients (P<0.05) (Table 2). Both the Kaplan-Meier model and the Cox proportional hazard regression model showed that MTERF3 expression was not significantly correlated with OS and DFS in breast cancer.
mRNA expression level of MTERF3 is correlated with other mitochondrial regulatory genes in breast cancer
The results of gene correlation analyses showed that the expression level of the MTERF3 gene in the breast cancer sample was positively correlated with that of the TFAM (P=1.4e−14, R=0.22), TFB1M (P=1e−11, R=0.2), TFB2M (P=0, R=0.34), MTERF1 (P=3.1e−08, R=0.16), TEFM (P=0, R=0.25) and MFN1 (P=2.5e−12, R=0.2) genes but was negatively correlated with that of MTERF4 (P=4.8e−07, R=−0.14) and PINK1 (P=1.4e−20, R=−0.26) genes. Meanwhile, there was no relevance between the MTERF3 and MTERF2 gene expression levels (P>0.05) (Figure 5).
Discussion
Breast cancer is the leading cause of cancer-related deaths worldwide, and its incidence continues to rise (22,23). Although the survival rate for breast cancer has improved significantly over the past 30 years as a result of advances in medical care and treatment, it is still the second leading cause of cancer-related deaths among women (24). Treatment failure and distant metastasis have always been the main challenges in breast cancer treatment. Therefore, it has become increasingly important to explore more markers to predict treatment response, tumor progression and potential therapeutic targets (25,26).
The MTERF family is a large protein family, that consists of four subfamilies (MTERF1-F4) that are widely expressed in metazoans and plants. The MTERF3 protein is the most evolutionarily conserved member and is a relatively primitive MTERF protein (27,28). MTERF3 contains five conserved mitochondrial transcription terminator motifs, including three leucine zippers (29). MTERF3 can inhibit transcription by binding to the promoter region of mtDNA, whereas MTERF3 knockout can increase the activity at the double-stranded mtDNA the transcription start site in cells. MTERF3 exerts a major inhibitory role on mtDNA expression and reduces the level of oxidative phosphorylation by decreasing the synthesis of respiratory enzymes, thereby reducing the generation of cellular energy (30). As reported in prior investigations, MTERF3 not only negatively regulates mtDNA transcription, but also modulates the assembly of large ribosome subunits, thus affecting ribosome biosynthesis and mitochondrial energy production (31). With in-depth research, there has been a growing realization that mitochondria play an important role in the occurrence and development of malignant tumors (32). Mitochondria are described as “cellular power plants”. Normal cells produce ATP by oxidative phosphorylation when there is sufficient oxygen to provide energy for cellular activity. However, decreased levels of oxidative phosphorylation in mitochondria may result in an imbalance between oxidative phosphorylation and glycolysis, which consequently induces the malignant transformation of cells (33) Mitochondria may also be closely associated with simultaneous abnormal energy metabolism, elevated reactive oxygen species (ROS), tissue infiltration and the metastasis of tumors (34,35). Compared to adjacent normal tissues, MTERF3 has been reported to be highly expressed in liver cancer, pancreatic cancer, lung cancer, breast cancer and brain glioma (16-18). In addition, Zhang et al. showed that up-regulation of MTERF3 gene could promote tumor growth both in vivo and in vitro, suggesting a role as a cellular oncogene (16). Notably, high expression of MTERF3 has been suggested to be positively correlated with tumor metastasis and invasion; estrogen or androgen independence and tumor immune resistance (16). The breast cancer cell MCF7 showed a higher MTERF3 expression in tamoxifen resistance cells than in tamoxifen sensitive cells (16). Previous studies have shown that mitochondrial gene mutation and changes in mitochondrial dynamics are closely associated with the occurrence and development of breast cancer (36). Nevertheless, as an important factor regulating mitochondrial gene expression, the correlation between the expression level of MTERF3 and clinicopathological features in breast cancer patients has not been reported.
In the present study, we investigated MTERF3 expression in breast cancer cells and breast cancer tissues. The protein level of MTERF3 was upregulated in breast cancer cell lines (BT-474, MCF7, SKBR3 and MDA-MB-468) compared with noncancerous breast cell (MCF10A). Strong MTERF3 staining was detected in breast cancer tissues, whereas absent or low expression was showed in most noncancerous breast tissues, which indicates that MTERF3 may display vital role in the carcinogenesis and progression of breast cancer. Our findings showed that MTERF3 was primarily located in the cytoplasm and the positivity rate of MTERF3 expression in breast cancer tissues was 91.38%, which was significantly higher than noncancerous breast tissues. Semiquantitative RT-PCR and western blot results showed that the mRNA and protein expression levels of MTERF3 in 58 breast cancer tissues were significantly increased compared to that in noncancerous breast tissues. We further evaluated the correlations between mRNA expression of MTERF3 and clinicopathological features as well as prognostic survival in breast cancer. Due to the relatively smaller number of breast cancer samples collected in the study, the R3.6.1 software was used to download the clinicopathological data of 1110 breast cancer patients with complete clinical data and the mRNA expression level of MTERF3 from the TCGA database to confirm the validity of the results. According to the analysis, high transcription level of MTERF3 was correlated with the ER status, PR status, breast cancer molecular typing, cancer type, histological diagnosis and primary site of breast cancer patients (P<0.05). No obvious correlation was observed with age, sex, tumor status, nodal status, metastasis status, AJCC stage, HER2 status or the history of other malignancies (P>0.05). These data suggested that the increased mRNA level of MTERF3 was closely related to the clinicopathological features of breast cancer patients. Kaplan-Meier analysis and log-rank test results showed that the mRNA expression level of MTERF3 was not correlated with OS or DFS in breast cancer. Subsequently, a Cox proportional risk regression model was used to analyze the prognosis of patients with breast cancer with univariate analysis and multivariate analysis. In the univariate analysis, factors such as age, tumor status, nodal status, metastasis status, AJCC stage, cancer type and the history of other malignancies influenced the prognosis of the patients. In the multivariate analysis, the age, metastasis status and cancer type of the breast cancer patients were independent factors leading to poor prognosis. However, there was no significant correlation between the expression level of MTERF3 and the prognosis of breast cancer patients. It is worth mentioning that another study has showed that breast cancer patients with lower MTERF3 level had a higher OS during the 200-months follow-up period (16). In the present study, we analyzed 705 breast cancer samples, the sample size was much larger and the results were more credible. Finally, gene correlation analysis showed that the TFAM, TFB1M, TFB2M, MTERF1, TEFM and MFN1 gene expression levels were positively correlated with MTERF3 expression, but MTERF3 expression was negatively correlated with MTERF4 and PINK1 gene expression. Based on the results, there was no correlation between the MTERF3 and MTERF2 gene expression levels. The results suggest that the role of MTERF3 in breast cancer is not independent, but interacts with other mitochondrial regulatory genes. The results of the present study provide insight into the roles of MTERF3 in breast cancer pathogenesis and suggest an association between mitochondrial gene transcription and breast cancer. However, this study only focuses on the bioinformatics analysis and pathology experiment, and these results may need additional cellular and molecular biology experiment to accurately identify the potential molecular mechanism.
Conclusions
In summary, this study demonstrated the differential expression of MTERF3 in noncancerous breast tissues and breast cancer tissues, suggesting that MTERF3 may be involved in the occurrence and development of breast carcinoma. Based on the analysis of the relationship of the expression level of MTERF3 with the clinicopathological characteristics and prognosis of breast cancer patients, MTERF3 cannot be used as a molecular marker for the prognosis of breast cancer patients. The findings of the present study may be beneficial for understanding the molecular biology mechanism of breast cancer and for generating effective therapeutic approaches for patients with breast cancer. As a newly discovered oncogene, the MTERF3 gene needs to be further studied to confirm its function and mechanism in human breast cancer.
Acknowledgments
Funding: This study was financially supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.65). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Dali University (No. 2014-12), and all patients signed an informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ma D, Chen C, Wu J, et al. Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and promotes carcinogenesis of breast cancer. Breast Cancer 2019;26:74-83. [Crossref] [PubMed]
- Winters S, Martin C, Murphy D, et al. Breast Cancer Epidemiology, Prevention, and Screening. Prog Mol Biol Transl Sci 2017;151:1-32. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012;486:405-9. [Crossref] [PubMed]
- Wagner BK, Kitami T, Gilbert TJ, et al. Large-scale chemical dissection of mitochondrial function. Nat Biotechnol 2008;26:343-51. [Crossref] [PubMed]
- Soriano M E, Scorrano L. Traveling Bax and Forth from Mitochondria to Control Apoptosis. Cell 2011;145:15-7. [Crossref] [PubMed]
- Hengartner M O. The biochemistry of apoptosis. Nature 2000;407:770-6. [Crossref] [PubMed]
- Heller A, Brockhoff G, Goepferich A. Targeting drugs to mitochondria. Eur J Pharm Biopharm 2012;82:1-18. [Crossref] [PubMed]
- Abramov AY, Smulders-Srinivasan TK, Kirby DM, et al. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain 2010;133:797-807. [Crossref] [PubMed]
- Weinberg S E, Chandel N S. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 2015;11:9-15. [Crossref] [PubMed]
- Fernandez-Silva P, Martinez-Azorin F, Micol V, et al. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J 1997;16:1066-79. [Crossref] [PubMed]
- Roberti M, Polosa PL, Bruni F, et al. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta 2009;1787:303-11.
- Roberti M, Polosa PL, Bruni F, et al. MTERF factors: a mutifunction protein family. Biomol Concepts 2010;1:215-24. [Crossref] [PubMed]
- Park CB, Asin-Cayuela J, Cámara Y, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 2007;130:273-85. [Crossref] [PubMed]
- Andersson DC, Fauconnier J, Park CB, et al. Enhanced cardiomyocyte Ca (2+) cycling precedes terminal AV-block in mitochondrial cardiomyopathy MTERF3 KO mice. Antioxid Redox Signal 2011;15:2455-64. [Crossref] [PubMed]
- Zhang CX, Wu N, Gao F, et al. MTERFD1 functions as an oncogene. Oncotarget 2016;8:1-8.
- Zi J, Wang W, Sun M, Mei W, et al. A high expression of MTERF3 correlates with tumor progression and predicts poor outcomes in patients with brain glioma. Int J Clin Exp Pathol 2019;12:1909-20. [PubMed]
- Sun S, Wu C, Yang C, et al. Prognostic roles of mitochondrial transcription termination factors in non-small cell lung cancer. Oncol Lett 2019;18:3453-62. [PubMed]
- Folescu R, Levai CM, Grigoraş ML, et al. Expression and significance of Ki-67 in lung cancer. Rom J Morphol Embryol 2018;59:227-33. [PubMed]
- Xiong W, Luo Y, Zhang C, et al. Expression, purification of recombinant human mitochondrial transcription termination factor 3 (hMTERF3) and preparation of polyclonal antibody against hMTERF3. Appl Biochem Biotechnol 2012;167:2318-29. [Crossref] [PubMed]
- Vuong D, Simpson PT, Green B, et al. Molecular classification of breast cancer. Virchows Arch 2014;465:1-14. [Crossref] [PubMed]
- Tsai WC, Chu CH, Yu CP, et al. Matriptase and Survivin Expression Associated with Tumor Progression and Malignant Potential in Breast Cancer of Chinese Women: Tissue Microarray Analysis of Immunostaining Scores with Clinicopathological Parameters. Disease Markers 2008;24:89-99. [Crossref] [PubMed]
- Sant M, Francisci S, Capocaccia R, et al. Time trends of breast cancer survival in Europe in relation to incidence and mortality. Int J Cancer 2006;119:2417-22. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Kabbage M, Trimeche M, Ben Nasr H, et al. Expression of the molecular chaperone alphaBcrystallin in infiltrating ductal breast carcinomas and the significance there of: an immunohistochemical and proteomics-based strategy. Tumour Biol 2012;33:2279-88. [Crossref] [PubMed]
- Elfagieh M, Abdalla F, Gliwan A, et al. Serum tumour markers as a diagnostic and prognostic tool in Libyan breast cancer. Tumour Biol 2012;33:2371-7. [Crossref] [PubMed]
- Linder T, Park CB, Asin-Cayuela J, et al. A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet 2005;48:265-9. [Crossref] [PubMed]
- Kleine T. Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front Plant Sci 2012;3:233. [Crossref] [PubMed]
- Spåhr H, Samuelsson T, Hällberg BM, et al. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid binding domain. Biochem Biophys Res Commun 2010;397:386-90. [Crossref] [PubMed]
- Roberti M, Bruni F, Loguercio Polosa P, et al. MTERF3, the most conserved member of the mTERF-family, is a modular factor involved in mitochondrial protein synthesis. Biochim Biophys Acta 2006;1757:1199-206.
- Wredenberg A, Lagouge M, Bratic A, et al. MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet 2013;9:e1003178. [Crossref] [PubMed]
- Kruspig B, Zhivotovsky B, Gogvadze V. Mitochondrial substrates in caner: drivers or passengers. Mitochondrion 2014;19 Pt A:8-19.
- Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S, et al. Modeling cancer glycolysis. Biochim Biophys Acta 2011;1807:755-67.
- Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, et al. Energy metabolism in tumor cells. FEBS J 2007;274:1393-418. [Crossref] [PubMed]
- Gupta SC, Hevia D, Patchva S, et al. Upsides and Downsides of Reactive Oxygen Species for Cancer: The Roles of Reactive Oxygen Species in Tumorigenesis, Prevention, and Therapy. Antioxid Redox Signal 2012;16:1295-322. [Crossref] [PubMed]
- Penta JS, Johnson FM, Wachsman JT, et al. Mitochondrial DNA in human malignancy. Mutat Res 2001;488:119-33. [Crossref] [PubMed]


