
The expression and the role of PIWI like RNA-mediated gene silencing 3 (PIWIL3) in lung cell line
Introduction
Lung cancer is the most common cancer, with 1.8 million new cases diagnosed annually, and is the leading cause of cancer-related deaths, with about 1.6 million deaths per year (1,2). Non-small cell lung cancers (NSCLC), including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, account for 80–85% of all lung cancers (2-4). Although there has been development in surgical resection, chemotherapy, and biologic treatment, the 5-year survival rate of only 18% is disconcerting, and is likely due to the high recurrence and metastasis of lung cancer (2,3). Therefore, understanding the underlying mechanism of recurrence and metastasis is critical to improving the outcome of treatment.
PIWI proteins, defined by highly conserved Piwi/Argonaute/Zwille (PAZ) and PIWI (P-element-induced wimpy testis) domains, form the core of the RNA-interference effector complex and RNA-induced silencing complexes (RISC) (5-7). PIWI family, including PIWIL1, PIWIL2, PIWIL3, and PIWIL4, has only thus far been identified in animals (8).
PIWIL3 involves in RNA silencing, translational regulation, and stem cell selfrenewal during the normal developmental process (9). Recently, PIWIL3 has been reported to be involved in the tumorigenesis and development of cancers (8,10,11). For example, RPS15A downregulation suppressed cell growth of gastric cancer (8) and breast cancer (10), while PIWIL3 overexpression inhibited glioma cell proliferation (11). However, the role of PIWIL3 in lung cancer is still unclear. Thus, we studied the role and function of PIWIL3 in lung cancer.
In this study, we first measured the expression of PIWIL3 in human lung cancer tissues. Then, we detected the expression of PIWIL3 in human lung cancer cell lines. Finally, we silenced PIWIL3 in A549 cells to determine the role of PIWIL3 in lung cancer cell proliferation, invasion, migration, and apoptosis.
Methods
Tissue samples
Thirty pairs of lung cancer and corresponding paracancer tissue samples were collected from lung cancer patients who underwent surgery in the First Affiliated Hospital of Bengbu Medical College from 2011 to 2012. None of the patients had previously undergone chemotherapy or radiotherapy. This study was approved by the local ethics committee of the First Affiliated Hospital of Bengbu Medical College, and the approval number is 2018KY005.
Immunohistochemistry
The immunohistochemistry using PIWIL3 antibody (1:300 dilution; abcam, Cambridge, United Kingdom; Cat. ab93709), followed by HRP-conjugated secondary antibody and DAB staining, was performed as described previously (12). The intensity of staining was recorded as 0 points (negative), 1 point (weak), 2 points (moderate), and 3 points (strong). The percentage of positive cells was accounted in 5 different high magnification views under a microscope, and was recorded as follows: 0 (no staining), 1 (<1/3 staining), 2 (1/3 to 2/3 staining), and 3 (>2/3 staining). A histoscore was calculated according to the intensity and percentage of positive cells (a score from 0 to 2 represents negative expression and a score >2 represents positive expression) (12,13).
Cell lines
Lung cancer cell lines A549, BEAS2, H1299, and H358 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), and were cultured in DMEM (Corning, Manassas, VA, USA) with 10% FBS (Corning, Manassas, VA, USA) and penicillin/streptomycin (37 °C, 5% CO2).
Knockdown PIWIL3 in A549 cells
siRNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) were used for infection in A549 cells based on the manufacturer’s instructions. The PIWIL3-specific interfering and non-silencing sequences were 5’-GCA ACT CAA AGA CCT GGA A-3’ and 5’-AAT TCT CCG AAC GTG TCA CGT-3’ (Sangon Biotech, Shanghai, China), respectively. Quantitative PCR (qPCR) was performed to measure infection efficiency.
The quantitative real-time PCR analysis
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA. The reverse transcription reaction was conducted based on the protocol of the M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). qPCR analysis was performed using the SYBR Master Mixture Kit (TaKaRa, Mountain, CA, USA) as described previously (14). GAPDH was applied as an internal control. The PIWIL3-specific primer sequences were 5’-GGATCAGCTACAACCCAGGAG-3’, and 5’-GTTCCTTCACCCCTTGAGACT-3’ (Sangon Biotech, Shanghai, China); the GAPDH-specific primer sequences were 5’-TGA CTT CAA CAG CGA CAC CCA-3’ and 5’-CAC CCT GTT GCT GTA GCC AAA-3’ (Sangon Biotech, Shanghai, China). The 2−ΔΔCT method was calculated to analyze the relative expression levels of PIWIL3.
Methylthiazoletetrazolium (MTT) proliferation assay
MTT assay was performed as previously described (15). In brief, at 72 hours after infection, A549 cells infected with shPIWIL3 (sh-PIWIL3 group) or shCtrl lentivirus (control group) were incubated with 15 µL of MTT reagent (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 4 hours. Then, 150 µL/well of dimethyl sulfoxide (DMSO; Sangon Biotech, Shanghai, China) was used to extract MTT formazan-formed crystal from cells. The amount of MTT formazan formed was determined by measuring the absorbance at 490 nm.
Colony formation assay
Cell colony formation was detected by a colony formation assay. A549 cells were infected with shPIWIL3 or shCtrl lentivirus at a density of 103/well. A549 cells were stained with crystal violet after infection for 10 days, and the number of colonies was counted under a microscope.
Apoptosis assay by fluorescence-activated cell sorting (FACS) analysis
Apoptosis assay was conducted as described previously (16). In brief, for apoptotic detection, exponentially growing A549 cells were seeded in 24-well plates. After infection with shPIWIL3 or shCtrl lentivirus for 3 days, cells were collected and resuspended with binding buffer. Then, the cells were incubated with 5 mL annexin V-APC (Thermo Fisher Scientific Inc, Carlsbad, CA, USA) for 10–15 minutes in the dark. After filtrating the cell suspension, the proportion of apoptotic cells was measured via FACS (Becton-Dickinson, Franklin Lakes, NJ, USA).
Transwell migration assay
Cell invasion was performed using a Transwell migration assay (17). Transwell chambers holding 8-µm pore polycarbonate membrane filters were used, with 2.5×103 cells cultured in each well. Trans-membrane A549 cells were dyed after infection si-PIWIL3 or shCtrol for 48 hours and observed under a microscope.
Wound scratch healing assay
Cell migration was conducted using a wound scratch healing assay (18). A549 cells were infected with shPIWIL3 or shCtrl lentivirus at a density of 2×104 cells/well in a 24-well plate. Cells were observed after infection for 48 hours.
Statistic analysis
All experiments were performed 3 replicates and the results were yielded based on the 3 replicates. Data were expressed as mean ± standard deviation (SD). Comparisons were performed by one-way ANOVA analysis and two-sided independent Student’s t-test. SPSS software version 20.0 (IBM SPSS Inc, Chicago, IL, USA) was used to analyze the data. Statistical significance was determined as a P value less than 0.05.
Results
The expression of PIWIL3 protein in lung cancer tissues
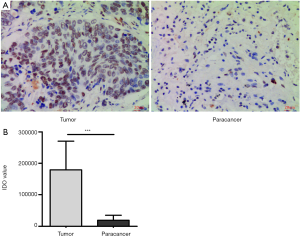
Immunohistochemistry was performed in 30 pairs of lung cancer and paracancer tissues to determine the expression of PIWIL3 in lung cancer tissues (Figure 1). The positive number of PIWIL3 in the lung cancer and paracancer tissues was 23 and 8, respectively. The positive rate of PIWIL3 in lung cancer tissues (70.7%) was significantly higher than that in paracancer tissues (26.7%). The IDO value was markedly higher in the tumor group than in the paracancer group (P<0.0001). Overall, we concluded that the expression of PIWIL3 protein is higher in lung cancer tissues compared to paracancer tissues.
The expression of PIWIL3 mRNA in lung cancer cell lines
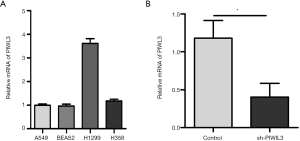
qRT-PCR was used to measure the expression of PIWIL3 mRNA in four lung cancer cell lines (A549, BEAS2, H1299, and H358). We found that PIWIL3 was expressed in all the four lung cancer cell lines (Figure 2A).
Knockdown PIWIL3 in A549 cells
To analyze the function of PIWIL3 in lung cancer, we silenced PIWIL3 in A549 cells by using siRNA and detected the infection efficiency using qRT-PCR at 48 hours after infection. As shown in Figure 2B, the expression of PIWIL3 mRNA was significantly decreased after using siRNA in A549 cells (P=0.0103).
PIWIL3 promotes A549 cell growth
To determine the effect of PIWIL3 on the proliferation of lung cancer cells, we measured the growth situation of si-PIWIL3-A549 and control-A549 cells. Firstly, the cell colony formation ability of si-PIWIL3-A549 and control-A549 cells was detected. As shown in Figure 3A, the number of A549 cells colonies dropped after silencing PIWIL3 in the image captured by the microscope. Also, we found that the number of colonies in the siPIWIL3-A549 group was markedly lower than that in the control-A549 group (P=0.0008) (Figure 3B). Furthermore, an MTT assay was conducted, and the OD490/fold ratio was recorded. As shown in Figure 3C, the OD490/fold ratio was significantly reduced in the siPIWIL3-A549 group compared to the control-A549 group at 72 hours after infection (P=0.0017) (Figure 3C).
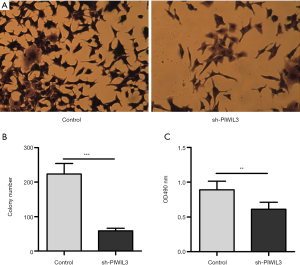
Overall, these results indicated that knockdown of PIWIL3 significantly inhibited the proliferation of lung cancer cell line A549.
PIWIL3 inhibits A549 cells apoptosis
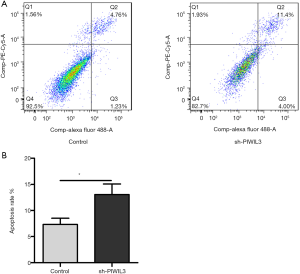
To analyze the effect of PIWIL3 on A549 cells apoptosis, we compared the apoptosis situation of siPIWIL3-A549 and control-A549 cells by flow cytometry. As shown in Figure 4A, the apoptosis rate in the siPIWIL3-A549 group was 11.4%, while that in the control-A549 group was only 4.76%. Also, by analyzing the results of flow cytometry, we found that the apoptosis rate of A549 cells increased dramatically after the treatment of siPIWIL3 (P=0.0140) (Figure 4B). Thus, we suggest that PIWIL3 can inhibit A549 cell apoptosis.
PIWIL3 promotes A549 cell invasion and migration
To investigate the effect of PIWIL3 on A549 cells invasion and migration, we conducted Transwell migration assay and wound scratch healing assay on siPIWIL3-A549 and control-A549 cells. As shown in the image of wound scratch healing assay obtained from microscopy (Figure 5A), and the result of cell counting (Figure 5B), the migration ability of A549 cells dropped significantly after silencing PIWIL3 (P=0.0004). Moreover, as shown in Figure 5C, we found that the number of trans-membrane A549 cells reduced after knocking down PIWIL3 in the image of Transwell migration assay. Also, we also found that the number of trans-membrane A549 cells in the siPIWIL3 group was markedly lower than that in the NC group (P=0.0004) (Figure 5D).
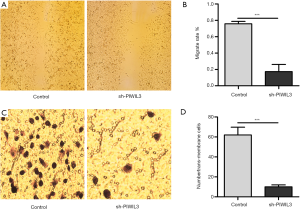
Overall, these results suggest that PIWIL3 can promote A549 invasion and migration.
Discussion
The PIWI family includes four isoforms of PIWI proteins, which are PIWIL1, PIWIL2, PIWIL3, and PIWIL4. Until now, the PIWI family could only be found in animals, while PIWIL3 was only identifiable in humans (8). PIWI genes exist in many organs, including the heart, lungs, kidney, liver, brain, and pancreas (19-23). PIWI genes are involved in RNA silencing, translational regulation, stem cell self-renewal, and spermatogenesis under physiological conditions (9,24). Also, the abnormal expression of PIWI genes can cause various pathological changes including the changing of lifespan and the dysregulation of fat metabolism (25). Recent studies show that the abnormal expression of PIWIL3 can cause the occurrence and progression of different tumors. However, the expression and the role of PIWIL3 in lung cancer is still unclear.
This study showed that PIWIL3 is highly expressed in lung cancer tissues and negatively related to the prognosis of lung cancer patients. Furthermore, PIWIL3 is widely expressed in lung cancer cells. Finally, silencing PIWIL3 can cause inhibition of growth and metastasis, as well as the promotion of apoptosis of A549 cells. Our research may furnish a useful and promising target for lung cancer treatment, and be a basis for further functional studies of PIWIL3 in lung cancer.
Previous studies have shown that PIWIL3 is highly expressed in colon cancer (9), gastric cancer (8), breast cancer (26), epithelial ovarian cancer (27), and melanoma tissues compared with corresponding normal tissues. For example, Li et al. (9) found that the positive rate of PIWIL3 in 75 colon cancer tissues was much higher than that in adjacent normal tissues by analyzing the results of a large tissue microarray. Also, Lim et al. (27) found that PIWIL3 was highly expressed in tumor and metastatic tumor tissues via detecting the expression of PIWIL3 in the primary tumor, paracancer, and peritoneal metastasis tissues, as well as in lymph nodes with or without metastasis. However, there exists inconsistent views concerning the expression of PIWIL3 in cancers (11,28). For instance, Iliev et al. (28) reported that there is no significant difference in the expression of PIEIL3 mRNA between 57 renal cell carcinoma tissues and paired renal parenchyma. Liu et al. (11) even found that the expression of PIWIL3 was lower in glioma tumor tissues compared to that in adjacent normal tissues, and its high expression was negatively related to tumor stage.
Further studies reported that the expression of PIWIL3 is related to tumor stage and the prognosis of patients. For example, high expression of PIWIL3 was reported to be positively related to the metastasis and thickness of melanoma (29), and negatively related to the overall survival rate of breast cancer patients (26). In this study, we measured the expression of PIWIL3 in lung cancer and adjacent normal tissues using immunohistochemistry and found that the positive rate of PIWIL3 in tumor tissues was higher than that in normal tissues.
The influence of PIWIL3 on both the biological functions of tumor cells and the situation of tumors in mouse models is also disputed (8,10,11). For example, Abell et al. (10) showed that after silencing PIWIL3, growth defects were found in breast cancer cells (MCF7). Also, Jiang et al. (8) concluded that the growth, invasion, and migration of gastric cancer cells, as well as the tumor volume in mice, were inhibited after knocking down PIWIL3. Conversely, it has been reported that the overexpression of PIWIL3 was found to inhibit cell proliferation and promote apoptosis of glioma cancer cells (U87 and U251), as well as induce tumor degeneration in mice (11). In this study, we compared the ability of growth, metastasis, and apoptosis in the lung cancer cells (A549) with and without PIWIL3 silencing, and found that PIWIL3 could promote the growth and metastasis of A549 cells, and inhibit the apoptosis of A549 cells.
Few studies have reported the biological mechanisms of PIWIL3. Jiang et al. (8) found that the downregulation of PIWIL3 in gastric cells (MKN45) could suppress the mRNA and protein levels of some metastasis-related genes which include ras homolog family member C (RhoC), metastasis-associated 1 (MTA1), matrix metallopeptidase 2 (MMP-2), and MMP-9. Also, they indicated that decreased PIWIL3 expression could block the activity of Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway by decreasing the phosphorylation levels of JAK2 and STAT3. As a long-chain non-coding gene, OIP5 antisense RNA 1 (OIP5-AS1) plays a biological role as an endogenous competition RNA (ceRNA) sponge of miR-367-3p (10). Liu et al. (10) reported that PIWIL3/OIP5-AS1/miR-367-3p/CCAAT enhancer-binding protein alpha (CEBPA) feedback loop regulates the biological behavior of glioma cells. These studies indicate a potential direction for studying the biological mechanisms of PIWIL3 in LC.
There are some limitations to this study. Firstly, only the PIWIL3 protein level was detected in lung cancer samples, and only PIWIL3 mRNA was detected in lung cancer cells in this study. However, PIWIL3 mRNA level in lung cancer samples and PIWIL3 protein level in lung cancer cells were not detected. Then, only one cell line was used to further analyze the function of PIWIL3 in lung cancer. Furthermore, we did not analyze the association of PIWIL3 with the clinical features of lung cancer patients. Finally, we did not perform experiments to explore potential biological mechanisms of PIWIL3 in LC. Hence, more related experiments and analyses should be done in further studies.
In conclusion, PIWIL3 was highly expressed in lung cancer and, our findings demonstrate that it can promote A549 cell growth and metastasis while inhibiting A549 cell apoptosis.
Acknowledgments
Thanks for all the staff in the Department of Oncology, The First Affiliated Hospital of Bengbu Medical College who have supported us.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.91). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the local ethics committee of the First Affiliated Hospital of Bengbu Medical College, and the approval number is 2018KY005. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 2018;22:3053-60. [PubMed]
- Skjefstad K, Johannessen C, Grindstad T, et al. A gender specific improved survival related to stromal miR-143 and miR-145 expression in non-small cell lung cancer. Sci Rep 2018;8:8549. [Crossref] [PubMed]
- Yu D, Qin Y, Jun-Qiang L, et al. CNPY2 enhances resistance to apoptosis induced by cisplatin via activation of NF-kappaB pathway in human non-small cell lung cancer. Biomed Pharmacother 2018;103:1658-63. [Crossref] [PubMed]
- Sun B, Liu HF, Ding Y, et al. Evaluating the diagnostic and prognostic value of serum miR-770 in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2018;22:3061-6. [PubMed]
- Chu H, Xia L, Qiu X, et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer 2015;121:2044-52. [Crossref] [PubMed]
- Fu A, Jacobs DI, Hoffman AE, et al. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 2015;36:1094-102. [Crossref] [PubMed]
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol 2007;3:36-43. [Crossref] [PubMed]
- Jiang L, Wang WJ, Li ZW, et al. Downregulation of Piwil3 suppresses cell proliferation, migration and invasion in gastric cancer. Cancer Biomarkers 2017;20:499-509. [Crossref] [PubMed]
- Li L, Yu C, Gao H, et al. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer 2010;10:38. [Crossref] [PubMed]
- Abell NS, Mercado M, Caneque T, et al. Click Quantitative Mass Spectrometry Identifies PIWIL3 as a Mechanistic Target of RNA Interference Activator Enoxacin in Cancer Cells. J Am Chem Soc 2017;139:1400-3. [Crossref] [PubMed]
- Liu X, Zheng J, Xue Y, et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics 2018;8:1084-105. [Crossref] [PubMed]
- Cao GD, Chen K, Chen B, et al. Positive prognostic value of HER2-HER3 co-expression and p-mTOR in gastric cancer patients. BMC Cancer 2017;17:841. [Crossref] [PubMed]
- Gulhati P, Cai Q, Li J, et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res 2009;15:7207-16. [Crossref] [PubMed]
- Fang KP, Dai W, Ren YH, et al. Both Talin-1 and Talin-2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC Cancer 2016;16:45. [Crossref] [PubMed]
- Moodley S, Koorbanally NA, Moodley T, et al. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay is a rapid, cheap, screening test for the in vitro anti-tuberculous activity of chalcones. J Microbiol Methods 2014;104:72-8. [Crossref] [PubMed]
- Juszczak K, Kaszuba-Zwoinska J, Thor PJ. Pulsating electromagnetic field stimulation of urothelial cells induces apoptosis and diminishes necrosis: new insight to magnetic therapy in urology. J Physiol Pharmacol 2012;63:397-401. [PubMed]
- Li W, Jia G, Qu Y, et al. Long Non-Coding RNA (LncRNA) HOXA11-AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial-Mesenchymal Transition. Med Sci Monit 2017;23:3393-403. [Crossref] [PubMed]
- Duan B, Guo T, Sun H, et al. miR-205 as a biological marker in non-small cell lung cancer. Biomed Pharmacother 2017;91:823-30. [Crossref] [PubMed]
- Henaoui I S, Jacovetti C, Guerra Mollet I, et al. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia 2017;60:1977-86. [Crossref] [PubMed]
- Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep 2015;5:10423. [Crossref] [PubMed]
- Rizzo F, Hashim A, Marchese G, et al. Timed regulation of P-element-induced wimpy testis-interacting RNA expression during rat liver regeneration. Hepatology 2014;60:798-806. [Crossref] [PubMed]
- Navarro A, Tejero R, Vinolas N, et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget 2015;6:31544-56. [Crossref] [PubMed]
- Nandi S, Chandramohan D, Fioriti L, et al. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc Natl Acad Sci U S A 2016;113:12697-702. [Crossref] [PubMed]
- Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006;442:203-7. [Crossref] [PubMed]
- Jones BC, Wood JG, Chang C, et al. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan. Nat Commun 2016;7:13856. [Crossref] [PubMed]
- Krishnan P, Ghosh S, Graham K, et al. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget 2016;7:37944-56. [Crossref] [PubMed]
- Lim SL, Ricciardelli C, Oehler MK, et al. Overexpression of piRNA pathway genes in epithelial ovarian cancer. PloS One 2014;9:e99687. [Crossref] [PubMed]
- Iliev R, Stanik M, Fedorko M, et al. Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients. Onco Targets Ther 2016;9:217-22. [PubMed]
- Gambichler T, Kohsik C, Hoh AK, et al. Expression of PIWIL3 in primary and metastatic melanoma. J Cancer Res Clin Oncol 2017;143:433-7. [Crossref] [PubMed]


