
Identification of key candidate genes and pathways in follicular variant papillary thyroid carcinoma by integrated bioinformatical analysis
Introduction
Thyroid carcinoma is the most common endocrine malignancy, accounting for ~2.1% of all cancer diagnoses worldwide, with ~77% of diagnoses occurrence in women, and the incidence has been increasing worldwide (1-4). The rate of new thyroid cancer cases has been raising by 3.8% on average each year for the last 10 years, while the death rate gained an average of 0.7% each year between 2005–2015. The 5-year survival trend ranged from 92.3% to 98.6% (5). Differentiated thyroid carcinoma (DTC), including papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC), account for 95% of thyroid cancer, and they have more favorable prognoses than poorly DTC (PDTC) and anaplastic thyroid cancer (ATC) (6). The most common variants of PTC include conventional PTC (classic/usual, cPTC), follicular variant PTC (FVPTC) and the tall cell variant PTC (TCPTC) (7).
FVPTC, one of the most frequent PTC variants (~30%), is a heterogeneous group of tumors that can be classified into two major groups, namely invasive/infiltrative FVPTC and noninvasive encapsulated FVPTC, which differ from each other in genes, clinic and morphology (8). The aggressive histopathologic features, such as thyroid capsule invasion, extrathyroidal extension and lymph node metastasis, are significantly less frequent in FVPTC than in cPTC, and the long-term outcome is either similar in both subtypes or more favorable in FVPTC compared with cPTC (9-11). Infiltrative FVPTC spreads to the lymph nodes, similar to cPTC, whereas encapsulated FVPTC behaves similar to the follicular adenoma (FA)/carcinoma group of tumors (12). Recently, the encapsulated/well demarcated non-invasive FVPTC was renamed “non-invasive follicular thyroid neoplasm with papillary-like nuclear feature” (NIFTP) by an international group of experts (12,13). FVPTC is less aggressive, so distinguishing between these two subtypes of PTC is necessary to avoid overtreatment and its complications. RNA sequencing provided a global view of the gene expression and enabled us to identify essential genes in the progression of thyroid carcinoma. Understanding the gene expression profiles of FVPTC as a whole will facilitate the research into the pathogenesis of this disease. However, the gene expression profiles, particularly the pathways in the interaction network and the diagnostic approaches of candidate markers of FVPTC, remain to be elucidated.
In this study, gene mutation and mRNA expression profiles of patients with FVPTC were obtained from The Cancer Genome Atlas (TCGA) database. Differentially expressed genes (DEGs) were identified, function enrichment and pathway analyses were performed, and then the protein-protein interaction (PPI) networks were constructed to identify the hub genes. We also performed receiver operating characteristic (ROC) analysis to evaluate the diagnostic values of the candidate genes. Our study may provide further insights into the investigation of the tumorigenesis mechanism of FVPTC and assist in the discovery of new candidate diagnostic markers for FVPTC.
Methods
TCGA data profiles
The TCGA thyroid carcinoma (TCGA-THCA) dataset consisted of 507 cases of PTC and 59 normal tissue samples (14). Merged level-3 RNA sequencing data and clinical information, including the BRAF V600E mutation, were downloaded from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/index.do) and Firebrowse (http://firebrowse.org). In total, 501 PTC cases and 59 normal samples with complete mRNA sequence and clinical data were identified. Among those 501 PTC cases, 357 cPTCs and 102 FVPTC (≥99% follicular patterned) cases were selected for inclusion in this study (a total of 459 sample IDs are shown in Tables S1,S2). RNAseq by expectation-maximization (RSEM) values was used to quantify mRNA expression levels. This study is exempt from ethics, as all data is derived from multiple public databases and that personal information is not identifiable.
Screening of DEGs
The differentially expressed mRNAs between the FVPTC and normal samples, between the cPTC and normal samples, and between the cPTC and FVPTC samples were analyzed with the Limma package (version 3.32.5) of R software (version 1.1.143). The function linear model fitting (lmFit) in the limma package was used to calculate fold changes (FC) and empirical Bayes statistics (eBayes) were used to estimate standard errors. Differentially expressed mRNAs with |logFC| >1.0 and adjusted P<0.05 were considered to indicate a statistically significant difference. Gene expression values were converted to log10 (RSEM) values using R. Hierarchical analysis of DEGs was achieved by Cluster 3.0 and then visualized via the Java TreeView 1.16r4 (http://www.treeview.net).
Functional enrichment analysis
Candidate DEG functions and pathways enrichment were carried out using the database for annotation, visualization and integrated discovery (DAVID, https://david.ncifcrf.gov/) online system. Gene ontology analysis (GO, classification including biological process, cellular component and molecular function) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment were performed. A P value <0.05 was set as the threshold.
PPI network analysis
The PPI network was constructed using the search tool for the retrieval of interacting genes/proteins (STRING, https://string-db.org, minimum required interaction score was set at 0.4) database and visualized with Cytoscape software 3.5.1. Sub-network models were selected using the plugin molecular complex detection (MCODE) application in Cytoscape. The criteria for defining a sub-network were an MCODE score ≥4 and the number of nodes ≥4 as described (15). Function enrichment of genes in sub-networks was performed using STRING. Hub genes were explored using the CytoHubba plugin application (16) by topological analysis of maximal clique centrality (MCC, top 10 nodes ranked by MCC).
Statistical analysis
Statistical analysis was carried out using SPSS 21.0 and R Studio (as described above). Chi-squared or Fisher’s exact test was used to analyze the association between different histological diagnoses and clinicopathological parameters. Survival analysis was analyzed using Kaplan-Meier and log-rank tests. ROC curves and the area under the curve (AUC) were performed to test the specificity and sensitivity of the candidate genes in predicting diagnosis (17). Cut-off values were determined by calculating the maximized sum of specificity and sensitivity. A P value of <0.05 and AUC >0.7 was considered to be statistically significant.
Results
Somatic mutation of FVPTCs and cPTCs
Gene mutation frequencies in the patients with PTC were obtained using cBioPortal. Furthermore, subgroup gene mutation studies of FVPTC and cPTC were queried by selecting relevant case IDs through the cBioPortal website. In total, altered genes (a mutation in more than 2 cases) happened in 78 of 102 sequenced FVPTC cases (Figure S1). The top 10 gene mutation profiles are shown in Figure 1. Alteration of RAS was particularly prevalent in FVPTC with a mutation frequency of 31% (including NRAS, HRAS, and KRAS mutations, the frequencies were 20%, 9%, and 2%, respectively). The BRAF mutation also took place at a high frequency (20%) in FVPTC. Distinguished with the FVPTC gene mutation pattern, the BRAF mutation was the predominant mutation type in cPTC (it occurred in 56% of cPTC patients), and attended by RET fusion (with a relatively low occurrence rate, at 9%). Notably, the incident rate of RAS mutation (including NRAS, HRAS, and KRAS mutations, the frequencies were 4%, 1.7%, and 0.71%, respectively) was much lower in the cPTC patients compared with the FVPTC patients.
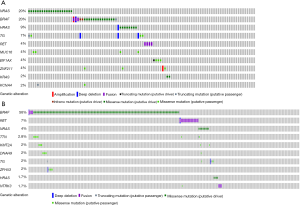
FVPTC seems to associate with favorable clinicopathological parameters
The correlation of two histological diagnoses with clinicopathological parameters was studied, to address the clinical differences of FVPTC and cPTC. The results showed that FVPTC was significantly correlated with the absence of extrathyroidal extension, lower N stage, and the low occurrence rate of BRAF mutation, while no significance was observed between patient gender, age, tumor focus type, T stage, and AJCC TNM stage (Table 1). Although a significant correlation (P=0.004) was observed between FVPTC and M1 stage, all of the PTC patients with distant metastasis were at a very low rate (9/507). Additionally, Kaplan-Meier analysis revealed no disease-free or overall survival rate difference between the patients with FVPTC or cPTC. These results suggested that FVPTC patients seem to present favorable clinicopathological features compared with the cPTC patients. Moreover, the FVPTC patients with a BRAF mutation significantly correlated with older age, extrathyroidal extension, and advanced TNM stage (Table S3).
Table 1
| Variables | No. of patients | cPTC | FVPTC | P value |
|---|---|---|---|---|
| Gender | 0.429 | |||
| Male | 122 | 98 | 24 | |
| Female | 337 | 259 | 78 | |
| Age | 0.396 | |||
| ≤45 | 224 | 178 | 46 | |
| >45 | 235 | 179 | 56 | |
| Multifocality | 0.198 | |||
| Unifocal | 253 | 202 | 51 | |
| Multifocal | 198 | 148 | 50 | |
| Extrathyroidal extension | <0.001* | |||
| None | 319 | 230 | 89 | |
| Minimal (T3) + moderate/advanced (T4a) | 123 | 111 | 12 | |
| T stage | 0.163 | |||
| T1 + T2 | 296 | 224 | 72 | |
| T3 + T4 | 161 | 131 | 30 | |
| N stage | <0.001* | |||
| N0 | 213 | 148 | 65 | |
| N1 | 197 | 184 | 13 | |
| M stage | 0.004** | |||
| M0 | 253 | 221 | 32 | |
| M1 | 9 | 4 | 5 | |
| AJCC TNM stage | 0.059 | |||
| Stage I + II | 318 | 240 | 78 | |
| Stage III + IV | 139 | 116 | 23 | |
| BRAF mutation | <0.001* | |||
| Without mutation | 239 | 157 | 82 | |
| BRAF V600E | 206 | 192 | 14 | |
| Disease free status | 0.363a | |||
| Disease free | 406 | 311 | 95 | |
| Recurred/progressed | 39 | 33 | 6 | |
| Overall survival status | 0.199a | |||
| Living | 443 | 342 | 101 | |
| Deceased | 16 | 15 | 1 |
Patients with censored data were excluded. *, P<0.05; **, P value by Fisher’s exact test; a, P value by log-rank test. FVPTC, follicular variant papillary thyroid carcinoma; cPTC, conventional papillary thyroid carcinoma; TCGA, The Cancer Genome Atlas.
Screening FVPTC-specific differentially expressed mRNAs
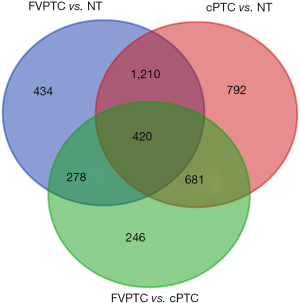
Differential analysis revealed 2,342 DEGs (1,695 downregulated and 647 upregulated genes) between the FVPTC and normal tissues (NT); 3,103 DEGs (1,723 downregulated and 1,380 upregulated genes) between the cPTC and NT with |logFC| >1.0 and adjusted P<0.05. DEGs between the FVPTC and NT were defined as FVPTC-related DEGs. Among these FVPTC-related DEGs, SLIT1, TMEM215, KLHDC8A, HAPLN1, and ZCCHC12 were the most significantly upregulated DEGs, while CCL21, DPT, MYOC, SCARA5, and RELN were the most significantly downregulated DEGs. Moreover, 1,625 DEGs were identified between FVPTC and cPTC with the same threshold, of which 1,191 were downregulated and 434 were upregulated. DEGs between FVPTC and cPTC that also belong to PTC-related DEGs (genes deregulated in both FVPTC and cPTC compared with NT) were defined as FVPTC-specific differentiated expressed genes (FVPTC-specific DEGs). According to this protocol, 420 FVPTC-specific DEGs, including 260 downregulated and 160 upregulated genes, were identified between the FVPTC and cPTC tissues (Figures 2,3). The average expression values in PTC tissues (including FVPTC and cPTC tissues); FC and P value details of these 420 deregulated genes are shown in Table S4. ZMAT4, SLC5A8, DIO1, MT1H, and CUX2 were the most significantly upregulated DEGs, while TMPRSS6, SFTPB, SYT12, SLC27A6, and TMPRSS4 were the most significantly downregulated DEGs in the FVPTC tissues compared with the cPTC tissues. Interestingly, the expression level of most oncogenes (PTC-related upregulated DEGs identified in our study) in the FVPTC tissues were much lower than in the cPTC tissues, while the expression level of most tumor suppressors (PTC-related downregulated DEGs) were higher in the FVPTC tissues than in the cPTC tissues (Table S4). These results suggested that tumor-related genes are less dysregulated in FVPTC compared with cPTC, which may partly explain why FVPTC is less aggressive than cPTC.
GO and KEGG pathway enrichment analysis of FVPTC-related and -specific DEGs
In order to gain a better understanding of the gene function and signaling pathways of FVPTC-related and -specific DEGs, online GO and KEGG pathway enrichment analysis were carried out using the DAVID system. For FVPTC-related DEGs, the top 5 enriched GO terms for biological process were cell adhesion, inflammatory response, immune response, cell-cell signaling, and signal transduction. The top 5 enriched GO terms for cellular component were integral component of plasma membrane, extracellular space, extracellular region, plasma membrane, and proteinaceous extracellular matrix. For molecular function heparin binding, calcium ion binding, chemokine activity, receptor activity, and Ras guanyl-nucleotide exchange factor activity were considerably enriched (Figure 4A). Furthermore, the enriched KEGG pathways of FVPTC-related DEGs included pathways in cancer, cytokine-cytokine receptor interactions, neuroactive ligand-receptor interactions, the PI3K-Akt signaling pathway, the MAPK signaling pathway, and the cell adhesion pathway (ranked by enriched gene number, Figure 4B).
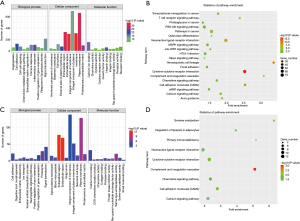
For FVPTC-specific DEGs, we found that the top 5 enriched GO terms for biological process were response to proteolysis, positive regulation of gene expression, keratan sulfate biosynthetic process, signal transduction, and cell adhesion. The top 5 enriched GO terms for cellular components were extracellular region, extracellular space, plasma membrane, proteinaceous extracellular matrix, and integral component of the plasma membrane. For molecular function, serine-type endopeptidase activity, serine-type peptidase activity, calcium ion binding, protein kinase inhibitor activity, and protease binding were significantly enriched (Figure 4C). Furthermore, those FVPTC-specific DEGs were significantly enriched in pathways in complement and coagulation cascades, tyrosine metabolism, cytokine-cytokine receptor interaction, cell adhesion molecules, and the chemokine signaling pathway (Figure 4D).
PPI network construction and Hub gene selection
In order to find the FVPTC-specific biomarkers, the PPI network was constructed only on 420 FVPTC-specific DEGs. By uploading the PPI network into the Cytoscape software, we found 228 nodules and 534 edges in the DEG network (Figure 5A). A total of 5 sub-networks were found using the definition criteria. The top sub-network, which included 12 nodules and 66 edges, was selected, and GO and KEGG pathway analyses were performed. Genes annotated in this sub- network included PYY, ADCY8, LPAR5, ADORA1, GRM4, CCL19, SSTR3, NPW, CCL21, CXCR5, C3, and NMU. The most significant enrichment terms were the G-protein coupled receptor signaling pathway, CCR7 chemokine receptor binding in GO, and the chemokine signaling pathway in KEGG (Figure 5B). The top 10 hub genes were annotated as LPAR5, NMU, CCL19, CCL21, C3, CXCR5, ADCY8, PYY, GRM4, and ADORA1 (ranked by MCC scores). All of these hub genes were included in the top sub-network. All of these results suggested that these hub genes might play an important role in the development of FVPTC.
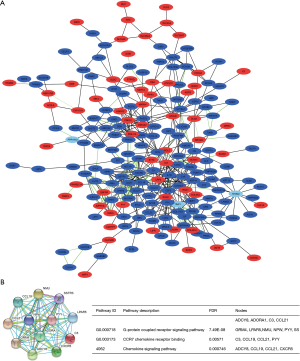
Diagnostic performance of hub genes
The diagnostic performance of the hub genes in differentiating FVPTC and cPTC from NT, and in differentiating FVPTC from cPTC (three independent groups) was evaluated by ROC analysis. To identify the discriminatory ability of these candidate genes in each group, AUC >0.7 was fixed as the threshold. First, we accessed the discriminatory ability of the hub genes between the FVPTC and NT. The upregulated genes ADCY8, ADORA1, and LPAR5 (Figure 6A) and the downregulated genes C3, CCL19, CCL21, CXCR5, and PYY (Figure 6B) were selected as the candidate genes with an AUC >0.7. Then, the discriminatory ability of the hub genes between the cPTC and NT was assessed. There were 8 hub genes, excluding CCL19 and CXCR5, possessed discriminatory abilities (Figure 6C,D). Additionally, C3 and ADORA1 exhibited the best prediction power with the highest AUCs (1 and 0.964, respectively). Finally, ROC analysis showed that only the downregulated genes (the genes downregulated in the FVPTCs compared with the cPTCs, which included ADCY8, ADORA1, C3, GRM4, LPAR5, and NUM) could robustly distinguish FVPTC from cPTC (Figure 6E). Furthermore, ADCY8, ADORA1, C3, and LPAR5 were identified in all of the three independent groups. The cut-off values and AUCs of these four genes for distinguishing the three independent groups are presented in Table 2. Collectively, our results suggest that combinatorial ADCY8, ADORA1, C3, and LPAR5 could not only be used for the differential diagnosis of PTC, including FVPTC and cPTC, from normal samples, but also for the differential diagnosis of FVPTC from cPTC samples.
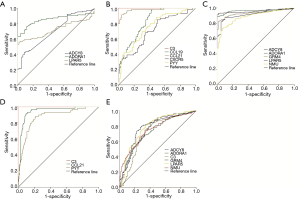
Table 2
| Candidate genes | Cut-off values (log2 RSEM) | AUC (sensitivity & specificity) | |||||
|---|---|---|---|---|---|---|---|
| FVPTC vs. NT | cPTC vs. NT | FVPTC vs. cPTC | FVPTC vs. NT | cPTC vs. NT | FVPTC vs. cPTC | ||
| ADCY8 | >0.631 | >0.317 | <2.421 | 0.704 (0.422, 0.915) | 0.920 (0.874, 0.898) | 0.774 (0.784, 0.725) | |
| ADORA1 | >7.740 | >8.404 | <9.246 | 0.852 (0.765, 0.881) | 0.964 (0.888, 1.000) | 0.724 (0.678, 0.676) | |
| C3 | <7.767 | <7.230 | <2.286 | 1.000 (1.000, 1.000) | 1.000 (1.000, 1.000) | 0.710 (0.748, 0.667) | |
| LPAR5 | >8.325 | >8.263 | <9.579 | 0.722 (0.559, 0.983) | 0.955 (0.930, 0.983) | 0.778 (0.810, 0.657) | |
NT, normal tissues; AUC, area under the curve; FVPTC, follicular variant papillary thyroid carcinoma; cPTC, conventional papillary thyroid carcinoma; RSEM, RNAseq by expectation-maximization.
Discussion
The gene mutation pattern and clinical characteristics of FVPTC were studied in the present study. Multi-group analyses were performed to identify aberrantly expressed genes in FVPTC. The FVPTC-related DEGs and FVPTC-specific DEGs were obtained from the TCGA-THCA dataset, and we proceeded with functional and pathway enrichment analysis. We highlighted the diagnostic values of several hub genes essentially, not only in distinguishing FVPTC and cPTC from NT, but also in discriminating FVPTCs from cPTCs. Through the TCGA-THCA data analysis, Stokowy et al. revealed molecular differences, including microRNA and mRNA, between FVPTC and cPTC. Putative miR-152-3p with the TGFA regulatory pair was discovered in FVPTC (18). Saiselet et al. illustrated that several downregulated miRNAs could distinguish histological classical variants and FVPTC in the TCGA dataset (19). To our knowledge, this is the first comprehensive study on gene mutation, screening FVPTC-specific DEGs, and the diagnostic approach for candidate biomarkers of FVPTC by utilizing TCGA-THCA data.
We found that more than half of the FVPTCs harbored oncogenic mutations, either the RAS (31%) or BRAF (20%) mutation. According to other reports, BRAF and RAS were the predominant mutations and mutually exclusive in all of the FVPTC cases (20). RAS mutations was demonstrated to get absolute predominance in encapsulated FVPTCs compared to infiltrative FVPTV (21). The incidence of the BRAF V600E mutation in encapsulated FVPTCs was relatively low, whereas an equal incidence of BRAF V600E or RAS mutation was observed in infiltrative FVPTCs (21,22). Yoo et al. revealed that the mutational profile of EFVPTC was similar to FA, while that of infiltrative FVPTC was similar to cPTC (23). We revealed that FVPTC seems to associate with favorable clinicopathological parameters compared with cPTCs, while FVPTC patients with the BRAF mutation significantly correlated with older age, extrathyroidal extension, and advanced TNM stage. Although the TCGA-THCA dataset did not provide the subgroup details of FVPTC, the FVPTC patients with the BRAF mutation appear to belong to a group with more aggressive behavior. Despite differences in the cytological classification and molecular profiles between noninvasive and infiltrative FVPTC, preoperative cytologic examination was indistinguishable between these two subgroups (21,22). Hence, diagnostic facilitation using molecular profiles combined with fine-needle aspirates (FNA) cytology should be evaluated by further studies with larger patient cohorts.
Slit guidance ligand 1 (SLIT1) belongs to the SLIT family of secreted proteins that mediate positional interactions between cells and their environment during development by signaling from ROBO receptors. SLITs and ROBOs are considered candidate tumor suppressor genes because their promoters are frequently hypermethylated in epithelial cancer (24). Hypermethylation of promotor region triggered SLIT1 downregulation, which has been reported in murine mammary glands or human breast carcinoma cells (24), glioma tumors (25), and early and advanced gastric cancers (26). PML knockdown led to impaired cell migration in glioblastoma, and the cell migration reduction was demonstrated to be associated with a marked upregulation of SLIT1 mRNA and protein levels, and of ROBO2. Furthermore, the PML/SLIT1 axis controls the sensitivity of glioblastoma cells to arsenic trioxide (27). There are only a few literature reviews the SLIT1 expression in thyroid normal and cancer tissues. Other than the downregulated expression pattern in other types of cancers, we found that SLIT1 was the top upregulated gene in FVPTC compared with the NT. Based on the above, the mechanism and function of upregulated SLIT1 in FVPTC need to be elucidated urgently. The C-C motif Chemokine ligand 21 (CCL21)/CCR7 axis promoted metastasis and survival of cancer cells by modulating EMT and cancer related pathways, and CCL21 correlated with tumor metastasis and prognosis (28,29). Lu et al. disclosed that CCL21 could facilitate chemoresistance and the stem cell property of colorectal cancer cells through AKT/GSK-3β/Snail signals (30). Zhang et al. disclosed that exogenous 100 ng/mL CCL21 markedly promoted cell proliferation, and CCL21/CCR7 interaction augmented the proportion of cells in G2/M via enhancing the p-ERK expression level (31). CCL21 was significantly lower in Ewing sarcoma patients with metastases than in patients without metastases. A higher RNA expression level of CCL21 was apparently corrected with good chemotherapeutic response and improved outcome (32). CCR7 was highly expressed in PTC (31), whereas the CCL21 expression level in PTC and the function of dysregulation of CCL21 should be elucidated. Hyaluronan- and proteoglycan-linked protein 1 (HAPLN1) showed essential effect on tumor proliferation, invasion, and metastasis. HAPLN1 is 23-fold overexpressed in stage 1 mesothelioma (33) and highly expressed in hepatocellular carcinomas (34) and oral cancer tissues (35). Zinc finger CCHC-type containing 12 (ZCCHC12) has important biological functions and acts as a metastasis-related oncogene in PTC (36). In our study, these cancer-related genes were also found to be dysregulated in FVPTCs; hence their function and mechanism in FVPTC need to be further investigated. Furthermore, the enriched KEGG pathways of FVPTC-related DEGs with the most genes were the pathways in cancer, the PI3K-Akt signaling pathway, the MAPK signaling pathway, and the cell adhesion pathway. Consistent with the results of other studies, the activation of the MAPK and PI3K pathways was observed in FVPTC (37). These data support that FVPTC shares the same pathway abnormalities with cPTC, although it has a different gene mutation pattern. Furthermore, the mild abnormality of molecular function, biological process, and related pathways involved in 420 FVPTC-specific DEGs may be one of the reasons for the lower malignant degree of FVPTC.
FNA cytology has the inherent limitation that indeterminate cytology results cannot distinguish between FA, FTC, or FVPTC. Molecular FNA diagnostics (combined FNA with molecular diagnostic approaches) is an essential complementary addition to FNA cytology (38). Previous studies have described the identification of biomarkers to improve the differential diagnosis of FVPTC from cPTC and benign lesions. Liu et al. identified that HBME-1 was profitable in the differentiation between FA and FVPTC (39). KAP-1, which was presented in 72% of FVPTC cases, could differentiate nodular hyperplasic lesions from both cPTC and FVPTC, while it was unable to distinguish FVPTC from FA and FTC (40). Hoftijzer et al. used a tissue array containing benign thyroid and thyroid carcinoma (including FVPTC) tissues to evaluate the diagnostic accuracy of RAR and RXR subtype protein expression for the differential diagnosis of thyroid neoplasms. In comparison between FA and FVPTC, nRARB, nRARA, cRXRB and mRXRB had sensitivities and specificities above 70%, and the highest specificity was for nRARB (91%) (41). Our study is the first to reveal that several biomarkers showed excellent predictive performances both in the differential diagnosis of PTC (including FVPTC and cPTC) from normal samples and in the differential diagnosis of FVPTC from cPTC samples. Future clinical studies with more cost-effective approaches, such as RT-PCR and immunohistochemical staining, instead of RNA sequencing should be conducted on a larger cohort of FVPTC samples to validate the clinical utility of the individual or combined hub genes identified in this study.
Conclusions
Our study identified the remarkable alterations of gene mutation, DEGs and related pathways in FVPTC. We demonstrated that the dysregulation of cancer-related genes might lead to diverse behavior in FVPTCs, even though the FVPTC shared the same pathway abnormalities with cPTC. In this study, we demonstrated 420 FVPTC-specific DEGs may be one of the reasons for the lower malignant degree of FVPTC. The diagnostic performance of hub genes may provide a novel mechanism in FVPTC and additional diagnostic targets of FVPTC. Differential diagnostic facilitation of FVPTC from cPTC and of FVPTC subgroups using molecular profiles should be evaluated by further studied with larger patients’ cohorts. More work is needed to elucidate the functional mechanism of these genes and to validate the clinical utility of these biomarkers with more cost-effective approaches.
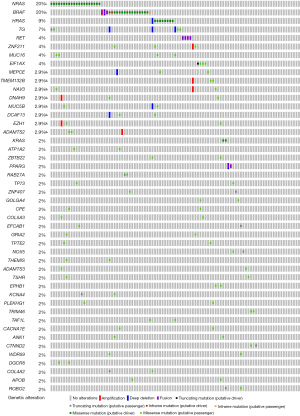
Table S1
| Sample IDs | Sample IDs | Sample IDs | Sample IDs |
|---|---|---|---|
| TCGA-4C-A93U | TCGA-DJ-A3US | TCGA-EL-A3T7 | TCGA-ET-A3BU |
| TCGA-BJ-A0YZ | TCGA-DJ-A3UU | TCGA-EL-A3T8 | TCGA-ET-A3BV |
| TCGA-BJ-A0Z0 | TCGA-DJ-A3UW | TCGA-EL-A3T9 | TCGA-ET-A3BW |
| TCGA-BJ-A0Z3 | TCGA-DJ-A3UX | TCGA-EL-A3TA | TCGA-ET-A3BX |
| TCGA-BJ-A0Z5 | TCGA-DJ-A3UY | TCGA-EL-A3TB | TCGA-ET-A3DO |
| TCGA-BJ-A0ZA | TCGA-DJ-A3UZ | TCGA-EL-A3ZG | TCGA-ET-A3DP |
| TCGA-BJ-A0ZB | TCGA-DJ-A3V2 | TCGA-EL-A3ZH | TCGA-ET-A3DR |
| TCGA-BJ-A0ZC | TCGA-DJ-A3V3 | TCGA-EL-A3ZK | TCGA-ET-A3DS |
| TCGA-BJ-A0ZE | TCGA-DJ-A3V9 | TCGA-EL-A3ZL | TCGA-ET-A3DT |
| TCGA-BJ-A0ZJ | TCGA-DJ-A3VA | TCGA-EL-A3ZM | TCGA-ET-A3DU |
| TCGA-BJ-A18Y | TCGA-DJ-A3VD | TCGA-EL-A3ZN | TCGA-ET-A40S |
| TCGA-BJ-A18Z | TCGA-DJ-A3VE | TCGA-EL-A3ZO | TCGA-ET-A40T |
| TCGA-BJ-A190 | TCGA-DJ-A3VI | TCGA-EL-A3ZP | TCGA-FE-A230 |
| TCGA-BJ-A192 | TCGA-DJ-A4UL | TCGA-EL-A3ZQ | TCGA-FE-A231 |
| TCGA-BJ-A28R | TCGA-DJ-A4UP | TCGA-EL-A3ZR | TCGA-FE-A232 |
| TCGA-BJ-A28S | TCGA-DJ-A4UT | TCGA-EL-A3ZS | TCGA-FE-A233 |
| TCGA-BJ-A28T | TCGA-DJ-A4V0 | TCGA-EL-A3ZT | TCGA-FE-A234 |
| TCGA-BJ-A28V | TCGA-DJ-A4V2 | TCGA-EL-A4JV | TCGA-FE-A235 |
| TCGA-BJ-A28W | TCGA-DJ-A4V5 | TCGA-EL-A4JW | TCGA-FE-A236 |
| TCGA-BJ-A28Z | TCGA-DO-A1K0 | TCGA-EL-A4JX | TCGA-FE-A237 |
| TCGA-BJ-A291 | TCGA-DO-A2HM | TCGA-EL-A4JZ | TCGA-FE-A238 |
| TCGA-BJ-A2N7 | TCGA-E3-A3E0 | TCGA-EL-A4K0 | TCGA-FE-A239 |
| TCGA-BJ-A2N8 | TCGA-E3-A3E1 | TCGA-EL-A4K1 | TCGA-FE-A23A |
| TCGA-BJ-A2N9 | TCGA-E3-A3E3 | TCGA-EL-A4K2 | TCGA-FE-A3PB |
| TCGA-BJ-A2NA | TCGA-E3-A3E5 | TCGA-EL-A4K4 | TCGA-FE-A3PC |
| TCGA-BJ-A2P4 | TCGA-E8-A242 | TCGA-EL-A4K6 | TCGA-FK-A3SB |
| TCGA-BJ-A3EZ | TCGA-E8-A2EA | TCGA-EL-A4K7 | TCGA-FK-A3SD |
| TCGA-BJ-A3F0 | TCGA-E8-A2JQ | TCGA-EL-A4K9 | TCGA-FK-A3SE |
| TCGA-BJ-A3PR | TCGA-E8-A3X7 | TCGA-EL-A4KD | TCGA-FK-A3SG |
| TCGA-BJ-A3PT | TCGA-E8-A413 | TCGA-EL-A4KG | TCGA-FK-A3SH |
| TCGA-BJ-A3PU | TCGA-E8-A414 | TCGA-EL-A4KH | TCGA-FK-A4UB |
| TCGA-BJ-A45C | TCGA-E8-A415 | TCGA-EL-A4KI | TCGA-FY-A2QD |
| TCGA-BJ-A45E | TCGA-E8-A416 | TCGA-EM-A1CS | TCGA-FY-A3BL |
| TCGA-BJ-A45F | TCGA-E8-A417 | TCGA-EM-A1CT | TCGA-FY-A3I4 |
| TCGA-BJ-A45H | TCGA-E8-A418 | TCGA-EM-A1CU | TCGA-FY-A3NM |
| TCGA-BJ-A45K | TCGA-E8-A419 | TCGA-EM-A1CV | TCGA-FY-A3NN |
| TCGA-BJ-A4O8 | TCGA-E8-A432 | TCGA-EM-A22I | TCGA-FY-A3ON |
| TCGA-CE-A13K | TCGA-E8-A433 | TCGA-EM-A22K | TCGA-FY-A3R6 |
| TCGA-CE-A27D | TCGA-E8-A434 | TCGA-EM-A22M | TCGA-FY-A3R7 |
| TCGA-CE-A3MD | TCGA-E8-A436 | TCGA-EM-A22O | TCGA-FY-A3R8 |
| TCGA-CE-A3ME | TCGA-E8-A437 | TCGA-EM-A22P | TCGA-FY-A3R9 |
| TCGA-CE-A481 | TCGA-E8-A438 | TCGA-EM-A2CS | TCGA-FY-A3RA |
| TCGA-CE-A482 | TCGA-E8-A44K | TCGA-EM-A2OX | TCGA-FY-A3TY |
| TCGA-CE-A483 | TCGA-E8-A44M | TCGA-EM-A2OZ | TCGA-FY-A3YR |
| TCGA-CE-A484 | TCGA-EL-A3CL | TCGA-EM-A2P0 | TCGA-FY-A40K |
| TCGA-CE-A485 | TCGA-EL-A3CM | TCGA-EM-A2P1 | TCGA-FY-A40L |
| TCGA-DE-A0XZ | TCGA-EL-A3CN | TCGA-EM-A2P3 | TCGA-FY-A40M |
| TCGA-DE-A0Y2 | TCGA-EL-A3CO | TCGA-EM-A3AK | TCGA-FY-A4B0 |
| TCGA-DE-A0Y3 | TCGA-EL-A3CR | TCGA-EM-A3AN | TCGA-FY-A4B3 |
| TCGA-DE-A3KN | TCGA-EL-A3CS | TCGA-EM-A3AO | TCGA-FY-A4B4 |
| TCGA-DE-A4M8 | TCGA-EL-A3CT | TCGA-EM-A3AQ | TCGA-FY-A76V |
| TCGA-DE-A4M9 | TCGA-EL-A3CU | TCGA-EM-A3AR | TCGA-GE-A2C6 |
| TCGA-DE-A4MA | TCGA-EL-A3CV | TCGA-EM-A3FJ | TCGA-H2-A26U |
| TCGA-DE-A4MB | TCGA-EL-A3CW | TCGA-EM-A3FK | TCGA-H2-A2K9 |
| TCGA-DE-A4MC | TCGA-EL-A3CX | TCGA-EM-A3FM | TCGA-H2-A3RI |
| TCGA-DE-A4MD | TCGA-EL-A3CY | TCGA-EM-A3FO | TCGA-H2-A421 |
| TCGA-DE-A69J | TCGA-EL-A3CZ | TCGA-EM-A3FQ | TCGA-H2-A422 |
| TCGA-DE-A69K | TCGA-EL-A3D0 | TCGA-EM-A3FR | TCGA-IM-A3EB |
| TCGA-DE-A7U5 | TCGA-EL-A3D1 | TCGA-EM-A3O3 | TCGA-IM-A3ED |
| TCGA-DJ-A13O | TCGA-EL-A3D4 | TCGA-EM-A3O7 | TCGA-IM-A420 |
| TCGA-DJ-A13P | TCGA-EL-A3D5 | TCGA-EM-A3SU | TCGA-J8-A3NZ |
| TCGA-DJ-A13T | TCGA-EL-A3D6 | TCGA-EM-A3SX | TCGA-J8-A3O0 |
| TCGA-DJ-A13U | TCGA-EL-A3GO | TCGA-EM-A3SZ | TCGA-J8-A3O1 |
| TCGA-DJ-A13V | TCGA-EL-A3GP | TCGA-EM-A4FF | TCGA-J8-A3O2 |
| TCGA-DJ-A1QD | TCGA-EL-A3GR | TCGA-EM-A4FM | TCGA-J8-A3YD |
| TCGA-DJ-A1QE | TCGA-EL-A3GS | TCGA-EM-A4FN | TCGA-J8-A3YE |
| TCGA-DJ-A1QF | TCGA-EL-A3GU | TCGA-EM-A4FO | TCGA-J8-A3YF |
| TCGA-DJ-A1QI | TCGA-EL-A3GV | TCGA-EM-A4FV | TCGA-J8-A3YG |
| TCGA-DJ-A1QN | TCGA-EL-A3GW | TCGA-ET-A25J | TCGA-J8-A3YH |
| TCGA-DJ-A1QQ | TCGA-EL-A3GX | TCGA-ET-A25K | TCGA-J8-A42S |
| TCGA-DJ-A2PN | TCGA-EL-A3GY | TCGA-ET-A25M | TCGA-J8-A4HW |
| TCGA-DJ-A2PO | TCGA-EL-A3GZ | TCGA-ET-A25N | TCGA-J8-A4HY |
| TCGA-DJ-A2PQ | TCGA-EL-A3H1 | TCGA-ET-A25O | TCGA-KS-A41F |
| TCGA-DJ-A2PR | TCGA-EL-A3H2 | TCGA-ET-A25P | TCGA-KS-A41J |
| TCGA-DJ-A2PS | TCGA-EL-A3H3 | TCGA-ET-A2MX | TCGA-KS-A4I1 |
| TCGA-DJ-A2PU | TCGA-EL-A3H4 | TCGA-ET-A2MY | TCGA-KS-A4I3 |
| TCGA-DJ-A2PV | TCGA-EL-A3H5 | TCGA-ET-A2MZ | TCGA-KS-A4I5 |
| TCGA-DJ-A2PW | TCGA-EL-A3H7 | TCGA-ET-A2N0 | TCGA-KS-A4I7 |
| TCGA-DJ-A2PZ | TCGA-EL-A3H8 | TCGA-ET-A39J | TCGA-KS-A4I9 |
| TCGA-DJ-A2Q0 | TCGA-EL-A3MW | TCGA-ET-A39K | TCGA-KS-A4IB |
| TCGA-DJ-A2Q1 | TCGA-EL-A3MX | TCGA-ET-A39L | TCGA-KS-A4IC |
| TCGA-DJ-A2Q4 | TCGA-EL-A3MY | TCGA-ET-A39M | TCGA-L6-A4EP |
| TCGA-DJ-A2Q5 | TCGA-EL-A3MZ | TCGA-ET-A39N | TCGA-L6-A4EQ |
| TCGA-DJ-A2Q6 | TCGA-EL-A3N2 | TCGA-ET-A39P | TCGA-MK-A4N6 |
| TCGA-DJ-A2Q7 | TCGA-EL-A3N3 | TCGA-ET-A39R | TCGA-MK-A4N7 |
| TCGA-DJ-A2QC | TCGA-EL-A3T0 | TCGA-ET-A39S | TCGA-MK-A4N9 |
| TCGA-DJ-A3UK | TCGA-EL-A3T1 | TCGA-ET-A3BN | TCGA-QD-A8IV |
| TCGA-DJ-A3UM | TCGA-EL-A3T2 | TCGA-ET-A3BP | – |
| TCGA-DJ-A3UN | TCGA-EL-A3T3 | TCGA-ET-A3BQ | – |
| TCGA-DJ-A3UO | TCGA-EL-A3T6 | TCGA-ET-A3BS | – |
cPTC, conventional papillary thyroid carcinoma; TCGA, The Cancer Genome Atlas.
Table S2
| Sample IDs | Sample IDs | Sample IDs | Sample IDs |
|---|---|---|---|
| TCGA-BJ-A0Z2 | TCGA-DJ-A3VM | TCGA-EM-A2CT | TCGA-ET-A25I |
| TCGA-BJ-A0ZG | TCGA-DJ-A4UR | TCGA-EM-A2CU | TCGA-ET-A2N3 |
| TCGA-BJ-A45D | TCGA-DO-A1JZ | TCGA-EM-A2OV | TCGA-ET-A2N4 |
| TCGA-BJ-A45G | TCGA-E3-A3DZ | TCGA-EM-A2OW | TCGA-ET-A2N5 |
| TCGA-BJ-A4O9 | TCGA-E3-A3E2 | TCGA-EM-A2OY | TCGA-ET-A39I |
| TCGA-DE-A2OL | TCGA-EL-A3CP | TCGA-EM-A2P2 | TCGA-ET-A39T |
| TCGA-DJ-A13M | TCGA-EL-A3GQ | TCGA-EM-A3AI | TCGA-ET-A3DQ |
| TCGA-DJ-A13R | TCGA-EM-A1CW | TCGA-EM-A3AJ | TCGA-ET-A3DV |
| TCGA-DJ-A13S | TCGA-EM-A1YA | TCGA-EM-A3AL | TCGA-ET-A40P |
| TCGA-DJ-A13W | TCGA-EM-A1YB | TCGA-EM-A3AP | TCGA-ET-A40R |
| TCGA-DJ-A13X | TCGA-EM-A1YC | TCGA-EM-A3FL | TCGA-ET-A4KQ |
| TCGA-DJ-A1QG | TCGA-EM-A1YD | TCGA-EM-A3FN | TCGA-FE-A22Z |
| TCGA-DJ-A1QL | TCGA-EM-A1YE | TCGA-EM-A3FP | TCGA-FE-A3PA |
| TCGA-DJ-A1QM | TCGA-EM-A22J | TCGA-EM-A3O6 | TCGA-FE-A3PD |
| TCGA-DJ-A2PP | TCGA-EM-A22L | TCGA-EM-A3O8 | TCGA-FK-A3S3 |
| TCGA-DJ-A2PX | TCGA-EM-A22N | TCGA-EM-A3O9 | TCGA-FY-A3I5 |
| TCGA-DJ-A2Q2 | TCGA-EM-A22Q | TCGA-EM-A3OA | TCGA-FY-A3NP |
| TCGA-DJ-A2QA | TCGA-EM-A2CJ | TCGA-EM-A3OB | TCGA-FY-A3W9 |
| TCGA-DJ-A2QB | TCGA-EM-A2CK | TCGA-EM-A3ST | TCGA-FY-A3WA |
| TCGA-DJ-A3UP | TCGA-EM-A2CL | TCGA-EM-A3SY | TCGA-FY-A40N |
| TCGA-DJ-A3UR | TCGA-EM-A2CN | TCGA-EM-A4FH | TCGA-H2-A3RH |
| TCGA-DJ-A3UT | TCGA-EM-A2CO | TCGA-EM-A4FK | TCGA-IM-A41Z |
| TCGA-DJ-A3VG | TCGA-EM-A2CP | TCGA-EM-A4FQ | TCGA-KS-A41I |
| TCGA-DJ-A3VK | TCGA-EM-A2CQ | TCGA-EM-A4FU | TCGA-KS-A41L |
| TCGA-DJ-A3VL | TCGA-EM-A2CR | TCGA-EM-A4G1 | TCGA-KS-A4ID |
| TCGA-L6-A4ET | – | – | – |
| TCGA-MK-A84Z | – | – | – |
FVPTC, follicular variant papillary thyroid carcinoma; TCGA, The Cancer Genome Atlas.
Table S3
| Variables | No. of patients | FVPTC with BRAF mutation | FVPTC without BRAF mutation | P value |
|---|---|---|---|---|
| Gender | 0.777 | |||
| Male | 24 | 4 | 20 | |
| Female | 78 | 16 | 62 | |
| Age | 0.049* | |||
| ≤45 | 46 | 5 | 41 | |
| >45 | 56 | 15 | 41 | |
| Multifocality | 0.625 | |||
| Unifocal | 51 | 9 | 42 | |
| Multifocal | 50 | 11 | 39 | |
| Extrathyroidal extension | 0.012* | |||
| None | 89 | 14 | 75 | |
| Minimal (T3) + moderate/advanced (T4a) | 12 | 6 | 6 | |
| T stage | 0.105 | |||
| T1 + T2 | 72 | 11 | 61 | |
| T3 + T4 | 30 | 9 | 21 | |
| N stage | 0.483 | |||
| N0 | 65 | 14 | 51 | |
| N1 | 13 | 4 | 9 | |
| M stage | 1.000 | |||
| M0 | 32 | 11 | 21 | |
| M1 | 5 | 1 | 4 | |
| AJCC TNM stage | 0.012* | |||
| Stage I + II | 78 | 10 | 68 | |
| Stage III + IV | 23 | 9 | 14 | |
| Disease free status | 0.442a | |||
| Disease free | 95 | 18 | 77 | |
| Recurred/progressed | 6 | 2 | 4 | |
| Overall survival status | 1.000a | |||
| Living | 101 | 20 | 81 | |
| Deceased | 1 | 0 | 1 |
Patients with censored data were excluded. P values were calculated by
Table S4
| Gene name | Average expression (log2 RSEM) | FVPTC vs. cPTC logFC | adj.P.Val | FVPTC vs. normal logFC | adj.P.Val | cPTC vs. normal logFC | adj.P.Val |
|---|---|---|---|---|---|---|---|
| ZMAT4 | 3.856925791 | 3.79867354 | 8.43E–21 | –2.086188414 | 2.50E–07 | –5.884861954 | 8.10E–32 |
| SLC5A8 | 5.763061203 | 3.489879204 | 1.26E–18 | –1.986582871 | 1.35E–06 | –5.476462075 | 2.11E–29 |
| DIO1 | 7.996888871 | 3.485550984 | 8.38E–17 | –1.475984973 | 0.001238834 | –4.961535956 | 6.33E–23 |
| MT1H | 5.016311976 | 3.451660858 | 5.91E–16 | –1.404487077 | 0.021328665 | –4.856147935 | 9.51E–24 |
| CUX2 | 2.686500534 | 3.421362768 | 9.35E–18 | –2.547930327 | 2.41E–05 | –5.969293094 | 4.77E–39 |
| TPO | 12.02223522 | 3.240204897 | 5.33E–19 | –1.710204819 | 3.05E–05 | –4.950409716 | 8.93E–30 |
| SELV | 2.951980747 | 3.07554214 | 3.18E–22 | –1.105874914 | 0.002838455 | –4.181417053 | 8.06E–29 |
| ZNF536 | 2.582310426 | 2.905251157 | 3.19E–22 | –1.368682963 | 8.41E–05 | –4.27393412 | 1.35E–32 |
| CARTPT | 2.514088207 | 2.857285915 | 1.12E–11 | –2.321984094 | 0.000623324 | –5.179270009 | 2.47E–28 |
| MRO | 4.55129724 | 2.850412043 | 4.82E–22 | –1.964071964 | 2.21E–09 | –4.814484006 | 1.67E–39 |
| WSCD2 | 5.724375037 | 2.816059629 | 4.13E–13 | –3.035673868 | 5.22E–13 | –5.851733498 | 5.27E–33 |
| STXBP5L | 4.723874543 | 2.810764377 | 7.90E–19 | –2.128601213 | 1.13E–09 | –4.939365591 | 1.24E–35 |
| CNTFR | 3.215658238 | 2.653743468 | 4.99E–12 | –1.241523575 | 0.030161691 | –3.895267043 | 8.67E–20 |
| RAG2 | 1.636336826 | 2.614759793 | 2.13E–13 | –3.166043678 | 1.99E–07 | –5.780803471 | 4.32E–46 |
| GATA5 | 2.622328675 | 2.56130456 | 5.90E–12 | 1.546839149 | 0.004209161 | –1.014465411 | 0.017956235 |
| HGD | 7.181194415 | 2.520110812 | 1.67E–23 | –1.179269118 | 1.95E–05 | –3.69937993 | 1.20E–33 |
| UPF0639 | 2.044555168 | 2.507920795 | 2.65E–20 | –1.41105203 | 2.82E–05 | –3.918972825 | 4.13E–33 |
| LRP1B | 3.87018084 | 2.475222817 | 5.00E–12 | –3.887786666 | 6.37E–16 | –6.363009483 | 3.75E–45 |
| TFF3 | 8.074375106 | 2.457307938 | 7.22E–13 | –4.048629329 | 2.85E–20 | –6.505937267 | 1.65E–48 |
| C12orf39 | 2.805268761 | 2.428203114 | 7.27E–21 | –1.539768701 | 3.02E–08 | –3.967971814 | 9.21E–34 |
| NEB | 5.232770027 | 2.399465335 | 1.29E–16 | –1.045270184 | 0.017068913 | –3.444735519 | 2.66E–24 |
| COL9A3 | 6.503023994 | 2.38528252 | 1.18E–11 | –1.153527361 | 0.049563902 | –3.538809881 | 1.36E–19 |
| ADIG | 1.837275909 | 2.372549768 | 2.57E–21 | –1.071849928 | 0.00117674 | –3.444399696 | 1.71E–31 |
| FABP4 | 5.604719077 | 2.355274638 | 1.48E–12 | –1.399428082 | 0.000469202 | –3.75470272 | 3.40E–21 |
| MPPED2 | 6.805085368 | 2.344361024 | 2.83E–22 | –1.6226023 | 9.72E–12 | –3.966963324 | 3.96E–38 |
| PKHD1L1 | 6.859408785 | 2.329195329 | 1.01E–07 | –3.663189107 | 1.61E–11 | –5.992384437 | 4.57E–29 |
| FLRT1 | 4.837889841 | 2.320044332 | 5.22E–19 | –1.257833709 | 6.61E–05 | –3.577878041 | 8.62E–30 |
| BMP8A | 8.430273753 | 2.307010208 | 2.59E–13 | –2.278471938 | 8.81E–07 | –4.585482146 | 3.14E–35 |
| GRIN2C | 6.797034664 | 2.29964708 | 7.25E–14 | –1.685246113 | 1.60E–06 | –3.984893193 | 1.11E–26 |
| C13orf36 | 3.581876173 | 2.287680021 | 8.14E–11 | –2.317003453 | 5.29E–06 | –4.604683474 | 2.28E–29 |
| SLC4A4 | 8.007270824 | 2.27684931 | 9.82E–22 | –1.540180775 | 1.62E–08 | –3.817030085 | 2.53E–38 |
| SEMA3D | 5.963454853 | 2.274904531 | 1.44E–09 | –3.230838782 | 2.29E–10 | –5.505743313 | 1.03E–33 |
| DPP6 | 5.069403151 | 2.243515517 | 6.89E–09 | –3.688950161 | 1.38E–14 | –5.932465678 | 3.66E–34 |
| EDN3 | 6.099794187 | 2.227574961 | 3.98E–08 | –1.63872395 | 0.000386931 | –3.866298911 | 1.87E–15 |
| LINGO2 | 2.079267748 | 2.19639063 | 2.78E–14 | –2.524003678 | 2.25E–10 | –4.720394308 | 4.47E–41 |
| NWD1 | 3.550766772 | 2.097507838 | 7.16E–13 | –2.244935631 | 6.89E–11 | –4.342443468 | 1.40E–32 |
| SH2D6 | 1.758658462 | 2.04853501 | 2.61E–17 | –1.915591863 | 4.27E–10 | –3.964126873 | 3.18E–40 |
| SLC26A4 | 10.8329384 | 2.021453084 | 3.78E–13 | –1.550136285 | 1.68E–07 | –3.571589369 | 2.34E–25 |
| GPR98 | 5.612700978 | 2.008564445 | 5.49E–16 | –1.939444746 | 1.16E–11 | –3.948009191 | 3.85E–36 |
| SLC26A7 | 11.00115332 | 1.991958092 | 1.17E–15 | –1.78099026 | 7.85E–15 | –3.772948351 | 7.87E–31 |
| TRPM3 | 1.419444405 | 1.986260032 | 1.06E–15 | –1.735209133 | 1.52E–06 | –3.721469165 | 4.51E–36 |
| MMEL1 | 1.143715589 | 1.976600748 | 2.32E–13 | –1.175592728 | 0.004969832 | –3.152193477 | 8.20E–26 |
| PPARGC1A | 8.012454456 | 1.902583916 | 8.12E–19 | –1.275211251 | 3.29E–07 | –3.177795167 | 1.50E–33 |
| MAPK4 | 5.010153953 | 1.891533755 | 6.02E–14 | –2.741672621 | 1.22E–13 | –4.633206377 | 1.51E–49 |
| DIRAS2 | 4.384236042 | 1.887947339 | 3.48E–20 | –1.431898963 | 4.70E–12 | –3.319846302 | 3.57E–36 |
| IGFBPL1 | 3.339526147 | 1.876866215 | 1.17E–12 | –2.62772169 | 7.14E–14 | –4.504587905 | 3.66E–42 |
| KCNA2 | 1.288454441 | 1.871972291 | 8.67E–17 | –1.05373759 | 0.002648886 | –2.925709882 | 5.05E–30 |
| KIAA1324 | 6.312086943 | 1.856665441 | 4.47E–11 | –2.045912421 | 3.59E–07 | –3.902577862 | 3.61E–32 |
| FAM189A1 | 5.28323919 | 1.852947373 | 2.48E–11 | –1.453996885 | 7.73E–07 | –3.306944258 | 1.71E–21 |
| TDGF1 | 2.05818749 | 1.851092704 | 1.27E–17 | –1.271409248 | 3.16E–06 | –3.122501952 | 1.68E–33 |
| HS6ST3 | 4.723290556 | 1.847056563 | 4.84E–13 | –1.591560226 | 4.71E–07 | –3.438616789 | 2.44E–28 |
| CWH43 | 4.699479584 | 1.838792946 | 2.43E–08 | –2.95439208 | 6.49E–14 | –4.793185026 | 7.26E–32 |
| GLDC | 3.742684454 | 1.796809029 | 1.14E–07 | –2.397391768 | 7.38E–06 | –4.194200797 | 1.87E–26 |
| PRIMA1 | 3.03785773 | 1.753697783 | 1.01E–10 | –1.578907864 | 9.04E–06 | –3.332605647 | 6.81E–26 |
| TPTE2P1 | 3.967440875 | 1.751888745 | 7.34E–11 | –2.000910967 | 4.28E–09 | –3.752799712 | 2.75E–29 |
| FER1L6 | 1.445594899 | 1.749990033 | 4.18E–09 | –3.037504007 | 3.01E–10 | –4.78749404 | 1.38E–38 |
| PKNOX2 | 6.212252026 | 1.749052111 | 6.46E–07 | –2.006883015 | 7.88E–07 | –3.755935126 | 3.23E–19 |
| CDH16 | 5.75779114 | 1.721825837 | 1.52E–06 | –4.081947241 | 1.14E–16 | –5.803773078 | 8.91E–40 |
| SOSTDC1 | 2.170469894 | 1.712559571 | 7.99E–10 | –1.771187511 | 7.54E–06 | –3.483747082 | 2.93E–26 |
| CYP17A1 | 3.959178957 | 1.709277151 | 1.73E–16 | –1.067413919 | 5.92E–05 | –2.776691069 | 2.13E–29 |
| PTPRQ | 2.58960269 | 1.707289088 | 8.99E–10 | –2.566740995 | 1.13E–10 | –4.274030083 | 7.60E–35 |
| NCAM1 | 8.703787505 | 1.701116354 | 1.10E–11 | –1.489161238 | 3.02E–07 | –3.190277592 | 7.13E–26 |
| PAX1 | 1.198048507 | 1.695690394 | 2.09E–09 | –2.664220545 | 1.30E–08 | –4.35991094 | 2.81E–40 |
| KLK15 | 0.800985115 | 1.690905779 | 2.30E–12 | –1.76908249 | 4.07E–05 | –3.459988269 | 1.73E–41 |
| TBX22 | 2.264680566 | 1.688774584 | 5.23E–09 | –3.069996666 | 1.74E–13 | –4.758771251 | 4.95E–39 |
| TFCP2L1 | 8.20717778 | 1.678503233 | 2.39E–14 | –2.455776252 | 2.41E–16 | –4.134279485 | 1.89E–47 |
| FHL1 | 11.43120601 | 1.641877191 | 1.61E–19 | –1.03649941 | 3.27E–08 | –2.678376601 | 4.37E–32 |
| MT1M | 5.35031493 | 1.606186773 | 8.67E–12 | –1.106387755 | 0.000373536 | –2.712574528 | 2.90E–22 |
| GRIK4 | 4.057163325 | 1.596925959 | 1.03E–09 | –1.671718171 | 5.82E–07 | –3.26864413 | 1.37E–24 |
| IRS4 | 2.153450243 | 1.596756793 | 9.80E–10 | –2.180716352 | 2.16E–07 | –3.777473145 | 2.94E–33 |
| SOD3 | 9.558384869 | 1.590393599 | 2.53E–18 | –1.005105997 | 4.82E–05 | –2.595499596 | 1.63E–33 |
| CRABP1 | 7.535983408 | 1.566634034 | 1.18E–05 | –2.738610074 | 1.32E–09 | –4.305244108 | 4.59E–24 |
| MIOX | 5.191718462 | 1.556241825 | 1.20E–10 | –1.422106327 | 1.54E–05 | –2.978348152 | 2.86E–26 |
| KIF19 | 3.497354052 | 1.552385999 | 2.03E–08 | –2.318228955 | 8.06E–11 | –3.870614954 | 1.55E–30 |
| C8orf12 | 0.69209291 | 1.536945879 | 6.89E–17 | –1.698650538 | 1.18E–07 | –3.235596418 | 1.11E–49 |
| FOXJ1 | 4.211993514 | 1.527911193 | 1.46E–06 | –2.540611324 | 6.95E–10 | –4.068522518 | 9.70E–26 |
| CA12 | 9.357862772 | 1.519496581 | 9.70E–09 | 2.954995202 | 7.99E–15 | 1.435498621 | 4.25E–06 |
| FOLR3 | 1.044288012 | 1.493990671 | 9.60E–12 | –1.511827835 | 1.44E–06 | –3.005818506 | 2.05E–30 |
| CIB4 | 1.371400165 | 1.472722781 | 5.90E–09 | –1.147896518 | 0.002413543 | –2.6206193 | 6.30E–20 |
| COL23A1 | 9.003120099 | 1.472541854 | 4.09E–11 | –1.693157251 | 1.13E–13 | –3.165699105 | 3.73E–29 |
| BEX1 | 6.509144008 | 1.463718771 | 5.48E–09 | –1.544118951 | 1.33E–07 | –3.007837723 | 1.86E–22 |
| ARHGAP24 | 8.295887083 | 1.460649534 | 3.55E–20 | –1.082300933 | 1.82E–09 | –2.542950467 | 3.55E–37 |
| ELMO1 | 9.276927183 | 1.455251171 | 4.26E–19 | –1.185728643 | 1.08E–08 | –2.640979814 | 8.51E–40 |
| HSD17B3 | 1.68670047 | 1.43028314 | 4.12E–10 | –1.927356647 | 1.65E–09 | –3.357639786 | 1.56E–33 |
| RFX4 | 0.609784334 | 1.417705733 | 1.34E–16 | –1.359189934 | 2.55E–09 | –2.776895667 | 5.05E–39 |
| CLCNKB | 6.51187325 | 1.409018078 | 2.46E–13 | –1.671497827 | 2.14E–10 | –3.080515905 | 1.93E–38 |
| CSMD1 | 4.056086398 | 1.404078414 | 3.64E–06 | 2.831382318 | 2.51E–10 | 1.427303904 | 3.13E–05 |
| FAM163A | 2.064494347 | 1.392929945 | 7.22E–09 | –2.462234887 | 2.31E–10 | –3.855164832 | 1.42E–37 |
| C6orf176 | 2.061954299 | 1.38228315 | 2.71E–06 | –2.246296818 | 3.32E–06 | –3.628579968 | 7.78E–23 |
| MUM1L1 | 6.320853527 | 1.365129184 | 2.39E–07 | –1.92748748 | 9.12E–10 | –3.292616663 | 1.85E–23 |
| TDRD9 | 5.813049876 | 1.361412223 | 1.67E–06 | –1.73175763 | 7.99E–06 | –3.093169853 | 2.31E–21 |
| SORBS2 | 11.00365408 | 1.356681195 | 1.04E–20 | –1.14223089 | 7.11E–10 | –2.498912085 | 5.06E–42 |
| FAM181B | 3.899716834 | 1.356099194 | 1.32E–13 | –1.035513473 | 1.40E–05 | –2.391612667 | 2.18E–27 |
| IPCEF1 | 7.608473519 | 1.348977751 | 8.74E–11 | –2.963237428 | 2.16E–18 | –4.31221518 | 1.33E–60 |
| DGKI | 5.385087185 | 1.346626571 | 8.95E–09 | –2.509231838 | 6.15E–14 | –3.855858409 | 7.79E–39 |
| TCEAL2 | 6.709267134 | 1.320042455 | 1.23E–05 | –2.292190972 | 3.63E–10 | –3.612233427 | 7.80E–23 |
| ASPG | 1.491397512 | 1.280607402 | 1.15E–08 | –1.727504814 | 1.02E–06 | –3.008112216 | 9.12E–31 |
| SYT9 | 3.085121255 | 1.275828769 | 1.23E–11 | –1.996235754 | 2.05E–13 | –3.272064522 | 1.40E–43 |
| ODZ2 | 3.116664704 | 1.266035166 | 3.57E–06 | –1.630455427 | 3.97E–06 | –2.896490594 | 9.49E–20 |
| ASXL3 | 2.592987042 | 1.261218652 | 5.39E–06 | –2.825024194 | 3.95E–12 | –4.086242845 | 2.20E–34 |
| NOL4 | 2.214207482 | 1.259914789 | 1.11E–05 | –3.259773686 | 6.42E–16 | –4.519688475 | 3.22E–37 |
| CD300LG | 2.458009472 | 1.258561482 | 6.03E–07 | –1.286252268 | 0.000192396 | –2.54481375 | 1.55E–17 |
| SAMD5 | 5.330023484 | 1.253865915 | 5.33E–12 | –1.081006312 | 0.000182436 | –2.334872227 | 2.56E–28 |
| C1orf64 | 0.897696717 | 1.251073938 | 1.59E–07 | –3.253510121 | 5.04E–17 | –4.504584059 | 1.78E–52 |
| WDR72 | 9.314943533 | 1.247027615 | 2.28E–13 | –1.513545963 | 1.35E–12 | –2.760573578 | 8.41E–38 |
| SH3GL2 | 3.095473839 | 1.246698113 | 2.71E–05 | –1.718762864 | 0.000696723 | –2.965460977 | 4.87E–20 |
| QRFPR | 3.645187179 | 1.244327481 | 2.85E–07 | 2.70169201 | 1.15E–16 | 1.457364529 | 6.99E–07 |
| ATP2C2 | 5.637482503 | 1.227483951 | 7.23E–08 | –2.680688568 | 2.51E–18 | –3.908172519 | 4.35E–42 |
| SLC29A4 | 6.654310934 | 1.218121293 | 3.11E–11 | –1.147157786 | 1.28E–05 | –2.365279079 | 2.09E–27 |
| KIAA1239 | 0.481487137 | 1.21306336 | 2.90E–08 | –2.613402422 | 2.01E–08 | –3.826465782 | 2.92E–54 |
| SLC24A3 | 5.726153247 | 1.208324196 | 1.59E–10 | –1.046810468 | 6.58E–06 | –2.255134664 | 1.77E–23 |
| KY | 6.371230296 | 1.206327432 | 2.34E–10 | –1.361999604 | 5.35E–10 | –2.568327036 | 1.79E–26 |
| ITPR1 | 9.669659374 | 1.201522627 | 6.22E–13 | –1.540928858 | 4.23E–14 | –2.742451485 | 7.35E–38 |
| GBA3 | 1.236439278 | 1.199064958 | 1.90E–07 | –1.467978937 | 1.41E–05 | –2.667043895 | 6.43E–25 |
| NXPH4 | 3.159523972 | 1.196939604 | 3.26E–10 | 2.63133703 | 1.35E–13 | 1.434397426 | 4.86E–12 |
| RAG1 | 4.779418513 | 1.194727341 | 3.55E–11 | –1.21425717 | 6.48E–06 | –2.408984512 | 1.92E–30 |
| AVPR1A | 4.147668499 | 1.177604299 | 1.37E–06 | –2.33180962 | 1.74E–12 | –3.509413918 | 3.35E–30 |
| ACSL6 | 1.608163103 | 1.169264798 | 6.55E–09 | –1.299470301 | 4.18E–05 | –2.468735099 | 1.89E–27 |
| PDE11A | 4.355202929 | 1.168068003 | 1.85E–07 | –1.724035822 | 9.11E–09 | –2.892103825 | 2.09E–27 |
| DLG2 | 6.458271635 | 1.166665087 | 3.35E–09 | –2.554377674 | 7.09E–18 | –3.721042761 | 1.40E–50 |
| COLEC10 | 2.416661893 | 1.166097884 | 1.94E–11 | –1.583129783 | 1.25E–10 | –2.749227667 | 2.50E–37 |
| TRIM50 | 1.195635056 | 1.165442553 | 5.25E–07 | –1.084357194 | 0.008483822 | –2.249799746 | 4.19E–20 |
| HKDC1 | 6.135824516 | 1.162421002 | 4.05E–07 | –1.334801089 | 4.64E–06 | –2.49722209 | 4.43E–20 |
| KIT | 8.192116399 | 1.156960126 | 1.29E–07 | –2.23741566 | 5.14E–13 | –3.394375786 | 1.06E–36 |
| RADIL | 3.871108248 | 1.153160002 | 5.08E–09 | –2.094732903 | 5.83E–16 | –3.247892905 | 1.89E–38 |
| RBM24 | 2.742612551 | 1.152759287 | 4.97E–10 | –1.56445767 | 1.27E–08 | –2.717216957 | 2.14E–33 |
| FOXP2 | 4.052486946 | 1.143341093 | 2.14E–05 | –2.369589525 | 1.16E–10 | –3.512930618 | 6.83E–26 |
| HAP1 | 4.336572237 | 1.138513092 | 1.11E–08 | –2.039146506 | 6.87E–11 | –3.177659598 | 5.58E–42 |
| UGT8 | 5.397699066 | 1.132318746 | 2.24E–08 | –1.11023398 | 8.88E–05 | –2.242552726 | 4.95E–21 |
| BRSK2 | 2.419522063 | 1.126252663 | 9.92E–12 | –1.018390681 | 0.000180933 | –2.144643344 | 1.07E–24 |
| C9orf24 | 1.514123909 | 1.124756805 | 1.77E–10 | –1.204861465 | 5.40E–06 | –2.32961827 | 4.18E–29 |
| PRKG1 | 5.256160513 | 1.116950858 | 9.29E–08 | –1.048452397 | 9.91E–05 | –2.165403255 | 1.85E–16 |
| LIFR | 9.16071807 | 1.115820295 | 3.12E–13 | –1.541844516 | 9.63E–12 | –2.657664811 | 8.04E–44 |
| HHIPL2 | 2.445617117 | 1.114723894 | 0.00049845 | –2.452875122 | 4.33E–07 | –3.567599016 | 1.03E–21 |
| LRP2 | 10.56563271 | 1.11462931 | 1.62E–07 | –1.195037249 | 1.92E–06 | –2.30966656 | 2.43E–19 |
| C3orf55 | 4.825184288 | 1.113423132 | 1.16E–08 | –1.349131721 | 5.39E–06 | –2.462554852 | 7.26E–29 |
| PCDP1 | 4.480423514 | 1.112748517 | 2.07E–12 | –1.257908764 | 1.54E–10 | –2.370657281 | 4.61E–33 |
| C11orf74 | 7.830416724 | 1.108077107 | 1.85E–25 | –1.107118012 | 8.36E–12 | –2.215195119 | 1.29E–61 |
| MAT1A | 2.144279497 | 1.09619335 | 4.59E–05 | –1.285180473 | 0.000714299 | –2.381373823 | 3.89E–15 |
| TMEM171 | 6.739583755 | 1.089905676 | 3.70E–11 | –1.887300124 | 1.70E–12 | –2.977205799 | 1.44E–49 |
| LRGUK | 1.508738041 | 1.081152781 | 1.43E–10 | –1.079432133 | 1.57E–05 | –2.160584914 | 3.33E–26 |
| SDK2 | 5.871863437 | 1.080107296 | 2.35E–09 | –1.130987196 | 7.85E–06 | –2.211094492 | 3.42E–25 |
| LRRC50 | 3.850296799 | 1.078632692 | 7.93E–09 | –1.800845571 | 4.80E–11 | –2.879478263 | 5.08E–37 |
| ODAM | 1.328438123 | 1.075434616 | 0.000109241 | –3.861547066 | 6.73E–19 | –4.936981682 | 1.71E–48 |
| FRMPD1 | 1.510383296 | 1.072988182 | 7.76E–09 | –1.147172989 | 8.58E–06 | –2.220161171 | 1.57E–23 |
| DEPDC6 | 9.720763342 | 1.071515398 | 2.65E–15 | –1.212222436 | 2.36E–12 | –2.283737835 | 1.02E–40 |
| ZNF804B | 2.222238243 | 1.059564117 | 0.002423893 | –4.687465038 | 7.55E–19 | –5.747029155 | 2.95E–40 |
| RNF150 | 5.105315074 | 1.05169778 | 2.53E–11 | –1.712263135 | 1.83E–12 | –2.763960915 | 3.23E–42 |
| FOXA2 | 4.590775464 | 1.050709906 | 0.000999837 | –2.471282168 | 6.40E–11 | –3.521992074 | 4.76E–20 |
| DCLK3 | 3.683484186 | 1.050392436 | 1.06E–10 | 2.455845219 | 8.88E–24 | 1.405452783 | 1.85E–12 |
| VEPH1 | 3.21881298 | 1.044745092 | 6.17E–10 | –1.181589247 | 3.86E–07 | –2.226334339 | 5.24E–28 |
| C8orf80 | 3.596175492 | 1.036419285 | 0.000472021 | –2.91157763 | 1.48E–16 | –3.947996915 | 3.81E–27 |
| TMEM90B | 5.474073991 | 1.034173818 | 6.14E–07 | –1.672932287 | 2.32E–09 | –2.707106106 | 2.36E–28 |
| PREX2 | 5.432350592 | 1.031741087 | 7.96E–07 | –1.204630326 | 2.77E–05 | –2.236371413 | 2.90E–19 |
| FMO1 | 2.521585809 | 1.030243875 | 1.90E–05 | –1.684068639 | 7.59E–09 | –2.714312514 | 5.40E–20 |
| PYY | 0.528408444 | 1.029658948 | 5.50E–07 | –1.989781506 | 3.48E–08 | –3.019440454 | 3.86E–30 |
| CNTN3 | 4.213727837 | 1.027448208 | 3.36E–05 | –1.451613403 | 8.29E–07 | –2.479061611 | 1.32E–16 |
| CNTNAP3 | 3.479498267 | 1.014504582 | 1.57E–08 | –1.338030661 | 1.60E–07 | –2.352535243 | 1.57E–27 |
| LRRC67 | 0.838436645 | 1.005868636 | 1.15E–09 | –1.087996435 | 8.58E–07 | –2.093865071 | 9.03E–25 |
| CNTN5 | 1.695903789 | 1.002825717 | 6.61E–05 | –3.795643069 | 3.50E–20 | –4.798468786 | 4.59E–49 |
| SLAMF6 | 4.853814775 | –1.001888147 | 4.89E–05 | –2.015375211 | 2.17E–07 | –1.013487064 | 0.000873935 |
| CHST2 | 10.82703068 | –1.002920532 | 3.89E–14 | 1.070476677 | 6.21E–08 | 2.073397209 | 5.17E–39 |
| GULP1 | 6.604415662 | –1.011855388 | 2.31E–08 | –2.3214821 | 6.68E–18 | –1.309626713 | 4.08E–10 |
| TSPAN11 | 5.5842067 | –1.014909968 | 1.31E–08 | –2.620027463 | 9.33E–22 | –1.605117495 | 2.78E–14 |
| SLC9A5 | 5.247910451 | –1.016984586 | 1.16E–13 | 1.005376365 | 1.19E–06 | 2.022360951 | 9.57E–35 |
| XG | 1.975143235 | –1.018233174 | 6.06E–06 | –2.663413021 | 9.52E–18 | –1.645179846 | 4.10E–09 |
| ADORA1 | 9.337751225 | –1.019711807 | 2.20E–12 | 1.581604986 | 5.75E–11 | 2.601316793 | 1.55E–50 |
| DCHS2 | 4.54149008 | –1.027660362 | 5.46E–07 | 1.090224238 | 0.000470506 | 2.1178846 | 1.23E–19 |
| APOBEC3H | 3.814850141 | –1.029011247 | 4.82E–10 | 1.036262078 | 0.000592148 | 2.065273325 | 6.33E–24 |
| GZMK | 4.518919563 | –1.031605051 | 0.000556172 | –2.337694453 | 3.40E–07 | –1.306089401 | 0.000309943 |
| CELF4 | 5.955547636 | –1.037262125 | 2.81E–08 | 1.950222607 | 4.16E–11 | 2.987484733 | 1.62E–41 |
| CSDC2 | 2.297873276 | –1.03810434 | 9.85E–06 | –2.464671377 | 6.19E–15 | –1.426567037 | 3.60E–07 |
| APOE | 12.55296554 | –1.038479232 | 2.64E–05 | 1.191306439 | 0.001567848 | 2.229785671 | 6.34E–15 |
| DUSP4 | 8.330060168 | –1.038596858 | 6.63E–15 | 1.533026974 | 3.41E–09 | 2.571623832 | 1.29E–47 |
| ADAM23 | 1.200189395 | –1.04316491 | 1.23E–06 | –2.793068561 | 8.79E–20 | –1.749903651 | 1.95E–11 |
| ALDH1A3 | 8.525595831 | –1.043517031 | 1.11E–05 | 1.551522636 | 4.17E–05 | 2.595039667 | 2.95E–22 |
| ZNF560 | 2.220201459 | –1.045698282 | 0.000752454 | 1.689544029 | 1.22E–06 | 2.735242311 | 7.24E–14 |
| PRELP | 8.114160947 | –1.050731443 | 6.81E–07 | –2.439457865 | 8.67E–15 | –1.388726422 | 6.85E–09 |
| SCML4 | 3.031729438 | –1.050743722 | 8.57E–07 | –2.270138334 | 1.11E–10 | –1.219394611 | 2.49E–06 |
| AMIGO2 | 9.126966805 | –1.05358616 | 3.90E–12 | 1.226307471 | 1.00E–07 | 2.279893631 | 7.71E–38 |
| CXCR5 | 2.197138086 | –1.055127147 | 0.000176951 | –2.321311754 | 2.31E–06 | –1.266184607 | 0.000866876 |
| ZNF385B | 1.55136712 | –1.06445282 | 2.91E–06 | –2.739641332 | 2.10E–17 | –1.675188512 | 2.48E–09 |
| KLHDC7B | 6.190927641 | –1.076232209 | 1.85E–05 | 1.253350891 | 0.004318282 | 2.3295831 | 7.80E–14 |
| PLA2G10 | 1.752475655 | –1.080096783 | 8.18E–05 | 1.501194523 | 1.67E–05 | 2.581291306 | 2.84E–16 |
| BTLA | 2.692219446 | –1.08248022 | 7.64E–06 | –2.090801291 | 1.93E–07 | –1.00832107 | 0.001212197 |
| PTGDS | 7.121108896 | –1.086535155 | 5.02E–05 | –2.571943076 | 6.41E–11 | –1.485407921 | 3.15E–06 |
| TMC6 | 10.5454448 | –1.086720311 | 4.61E–19 | 1.50357252 | 2.57E–11 | 2.59029283 | 2.40E–54 |
| OLAH | 3.850654515 | –1.088878741 | 2.42E–05 | 2.252997947 | 7.53E–11 | 3.341876688 | 3.15E–26 |
| B3GNT7 | 8.610471571 | –1.089648249 | 3.43E–11 | 1.359198034 | 5.01E–06 | 2.448846283 | 2.83E–45 |
| PTGER1 | 1.743015123 | –1.096285415 | 1.04E–07 | 1.279031191 | 1.22E–06 | 2.375316605 | 1.31E–21 |
| FAM111B | 6.464614128 | –1.096914809 | 1.57E–10 | 1.744995487 | 1.21E–09 | 2.841910295 | 2.76E–34 |
| SERPINF1 | 8.794692221 | –1.103280845 | 1.77E–09 | –2.30151395 | 1.09E–19 | –1.198233104 | 2.71E–08 |
| KLK13 | 4.331270536 | –1.1057771 | 1.60E–07 | 2.258207412 | 3.87E–10 | 3.363984512 | 1.27E–39 |
| CCL21 | 5.214233905 | –1.106469562 | 0.025479573 | –7.487311461 | 1.46E–27 | –6.380841899 | 4.33E–28 |
| CES1 | 6.137003008 | –1.107011149 | 0.000896459 | –2.725807152 | 4.46E–07 | –1.618796003 | 4.08E–06 |
| LRP4 | 11.62056967 | –1.108440735 | 3.53E–08 | 3.217320539 | 2.49E–19 | 4.325761273 | 4.64E–66 |
| LAD1 | 9.557167768 | –1.120986738 | 4.86E–15 | 1.293886656 | 1.90E–11 | 2.414873395 | 7.78E–42 |
| SLC43A3 | 10.15733088 | –1.126890987 | 1.35E–11 | –2.332862468 | 6.26E–23 | –1.20597148 | 6.56E–10 |
| ALK | 4.987852581 | –1.127085788 | 1.50E–07 | 1.964012709 | 1.47E–11 | 3.091098496 | 2.27E–32 |
| GDF15 | 10.17672836 | –1.127406362 | 3.49E–07 | 3.112439747 | 1.26E–17 | 4.239846109 | 4.42E–49 |
| UPP1 | 9.549698491 | –1.129773889 | 3.56E–19 | 1.018325905 | 1.87E–10 | 2.148099794 | 5.65E–39 |
| ITK | 4.84600987 | –1.131929848 | 1.36E–06 | –2.329449503 | 1.21E–10 | –1.197519655 | 3.86E–05 |
| PTGIS | 5.598486417 | –1.137027894 | 1.61E–06 | –2.511873059 | 5.26E–15 | –1.374845165 | 8.87E–07 |
| RAET1E | 3.927380278 | –1.141187192 | 9.32E–11 | 1.61824012 | 6.07E–13 | 2.759427312 | 5.05E–33 |
| SHROOM4 | 10.26125297 | –1.149615489 | 1.60E–22 | 1.609101411 | 1.63E–21 | 2.7587169 | 1.19E–65 |
| BID | 9.20751749 | –1.163930631 | 4.08E–26 | 1.145893761 | 1.23E–11 | 2.309824393 | 1.05E–57 |
| BHLHE22 | 2.017723353 | –1.16460548 | 3.73E–07 | –3.199883701 | 7.97E–26 | –2.035278221 | 1.46E–13 |
| CD40LG | 2.707240961 | –1.165971589 | 6.55E–07 | –2.682163555 | 7.13E–14 | –1.516191966 | 1.29E–07 |
| OGN | 4.331256873 | –1.166653157 | 0.001178506 | –5.184833284 | 2.62E–23 | –4.018180128 | 2.26E–21 |
| DPT | 5.055874789 | –1.166742915 | 0.002830981 | –6.580477797 | 1.37E–29 | –5.413734882 | 8.26E–32 |
| MPZL2 | 12.18676894 | –1.169702341 | 4.56E–25 | 1.236912803 | 2.02E–13 | 2.406615144 | 1.06E–63 |
| OSR1 | 3.052189343 | –1.17202349 | 9.66E–08 | –4.13367286 | 6.53E–30 | –2.96164937 | 9.86E–29 |
| PROM1 | 3.623389776 | –1.175587625 | 0.00029696 | –4.963015599 | 3.68E–23 | –3.787427974 | 3.92E–22 |
| KISS1R | 2.649819535 | –1.176514313 | 4.56E–09 | 1.978626964 | 1.68E–11 | 3.155141277 | 2.85E–37 |
| EGFL6 | 2.975502896 | –1.176944161 | 7.23E–07 | 1.146178902 | 0.000851509 | 2.323123064 | 1.93E–16 |
| GALE | 10.15875429 | –1.179276518 | 3.10E–16 | 1.895567425 | 1.07E–14 | 3.074843942 | 8.20E–69 |
| CDKN2A | 5.369805922 | –1.191174742 | 1.75E–09 | 1.859893056 | 1.05E–11 | 3.051067799 | 2.12E–31 |
| FST | 2.363707928 | –1.202112076 | 4.88E–07 | –2.675821732 | 5.12E–19 | –1.473709657 | 2.10E–07 |
| CA7 | 3.200035712 | –1.209876147 | 2.74E–08 | 1.582359019 | 3.29E–08 | 2.792235166 | 1.23E–25 |
| TIAM1 | 9.352406209 | –1.211818781 | 9.81E–11 | 1.86124259 | 3.29E–10 | 3.073061372 | 6.69E–41 |
| TMEM90A | 5.202964143 | –1.212595474 | 6.37E–09 | 2.757607803 | 4.65E–13 | 3.970203278 | 8.16E–54 |
| MSC | 4.564391708 | –1.213785604 | 3.27E–09 | –2.270561163 | 1.77E–14 | –1.056775559 | 1.52E–05 |
| KIAA1045 | 2.422733488 | –1.22123972 | 5.32E–12 | –2.687025328 | 3.74E–22 | –1.465785608 | 3.66E–12 |
| LY9 | 3.673018842 | –1.225839462 | 2.62E–06 | –2.299951658 | 1.11E–07 | –1.074112197 | 0.000881624 |
| METTL7B | 9.933248532 | –1.226736813 | 1.83E–11 | 2.988394058 | 3.92E–18 | 4.215130871 | 5.68E–77 |
| KRT14 | 3.164245511 | –1.227125868 | 7.88E–05 | 1.163954473 | 0.008415461 | 2.391080342 | 2.67E–10 |
| FAM132A | 3.396880016 | –1.227281047 | 3.25E–09 | 1.774441794 | 1.02E–09 | 3.001722841 | 8.05E–31 |
| CITED1 | 10.96254457 | –1.230524099 | 4.23E–05 | 3.400126387 | 5.64E–13 | 4.630650486 | 6.96E–39 |
| MYOC | 2.162454826 | –1.233545154 | 0.000289963 | –6.578094747 | 1.03E–32 | –5.344549593 | 2.78E–35 |
| PODN | 6.271775108 | –1.234090053 | 1.76E–07 | –4.010806545 | 4.09E–27 | –2.776716492 | 5.03E–23 |
| CDH4 | 5.253811381 | –1.244523905 | 2.71E–05 | 2.185717894 | 3.29E–07 | 3.430241799 | 1.16E–22 |
| TUSC3 | 9.628227631 | –1.245875488 | 5.30E–13 | 1.920820511 | 1.19E–10 | 3.166695999 | 9.95E–54 |
| CD3G | 2.962773363 | –1.247281058 | 4.61E–07 | –2.410797029 | 1.17E–10 | –1.163515971 | 0.000133145 |
| C8orf31 | 5.42733817 | –1.249123316 | 2.81E–11 | –2.384714336 | 8.92E–17 | –1.13559102 | 5.44E–08 |
| ODZ3 | 7.806195395 | –1.26772812 | 3.08E–08 | –2.778880319 | 1.64E–14 | –1.511152199 | 1.15E–09 |
| PNMT | 4.399487805 | –1.273568429 | 2.88E–08 | 1.503982755 | 0.000213052 | 2.777551184 | 1.92E–28 |
| AGR2 | 9.100664163 | –1.278642899 | 7.99E–06 | 2.361508382 | 1.17E–05 | 3.640151281 | 3.09E–34 |
| ENTPD8 | 5.311315302 | –1.285947308 | 2.97E–11 | 1.328960328 | 2.56E–05 | 2.614907636 | 4.29E–31 |
| PDPN | 5.14088594 | –1.28915272 | 2.11E–09 | –2.575684151 | 1.12E–14 | –1.286531431 | 1.41E–07 |
| CHRDL1 | 5.101670588 | –1.295692467 | 0.000196353 | –5.970964318 | 6.37E–28 | –4.675271851 | 1.32E–29 |
| HIST3H2A | 3.580321216 | –1.298519219 | 3.19E–07 | 1.077662436 | 0.005800695 | 2.376181655 | 9.67E–17 |
| GOLT1A | 7.412787975 | –1.299161922 | 4.71E–11 | 2.792181627 | 3.55E–13 | 4.091343549 | 4.69E–71 |
| DUSP6 | 13.00753383 | –1.30410373 | 3.49E–20 | 1.128994136 | 3.86E–07 | 2.433097866 | 1.41E–44 |
| COL17A1 | 7.284263821 | –1.308782421 | 4.53E–06 | –2.55339586 | 7.26E–11 | –1.244613439 | 0.000120322 |
| HRH1 | 7.621377527 | –1.313928187 | 2.59E–19 | 1.165155575 | 4.20E–13 | 2.479083762 | 9.84E–38 |
| C1R | 9.677396383 | –1.317298932 | 7.74E–12 | –2.681081123 | 5.54E–23 | –1.363782191 | 2.19E–09 |
| FREM1 | 1.670501657 | –1.317520159 | 5.55E–08 | –3.773449282 | 2.12E–31 | –2.455929123 | 3.25E–16 |
| PROC | 3.415997722 | –1.336060869 | 9.09E–12 | 1.13370471 | 0.000101649 | 2.469765579 | 8.13E–25 |
| RIN1 | 7.099458254 | –1.339121071 | 8.01E–25 | 1.103204854 | 5.54E–09 | 2.442325925 | 1.79E–50 |
| CTSH | 12.43737375 | –1.341142074 | 1.57E–17 | 1.052253061 | 3.56E–06 | 2.393395135 | 2.92E–36 |
| GGT6 | 5.923735296 | –1.342855363 | 9.56E–06 | 1.403673088 | 0.006776951 | 2.746528451 | 9.36E–18 |
| MMP11 | 7.465578309 | –1.345309462 | 1.26E–11 | 1.597067297 | 8.63E–12 | 2.942376759 | 8.64E–31 |
| GREM1 | 4.514014862 | –1.35356223 | 1.29E–05 | –2.459922452 | 4.04E–09 | –1.106360223 | 0.002381091 |
| LRRC52 | 2.413635904 | –1.362740821 | 3.65E–12 | 1.173272994 | 2.71E–05 | 2.536013815 | 1.03E–26 |
| CDH6 | 8.374845725 | –1.363388437 | 2.33E–17 | 1.413901439 | 1.14E–15 | 2.777289876 | 2.87E–39 |
| LOC153328 | 1.675438505 | –1.365287763 | 1.15E–09 | 1.013130142 | 7.39E–05 | 2.378417905 | 2.11E–18 |
| TRIM63 | 3.487696083 | –1.372109078 | 4.95E–07 | 1.642352808 | 0.000224012 | 3.014461886 | 5.32E–21 |
| C1orf170 | 5.555284475 | –1.372205596 | 3.34E–15 | 2.132758626 | 4.18E–13 | 3.504964222 | 1.75E–53 |
| GSDMA | 3.754032329 | –1.390361658 | 3.79E–10 | 1.342138281 | 4.31E–05 | 2.732499938 | 2.02E–23 |
| NALCN | 4.860185588 | –1.39581601 | 5.37E–07 | –2.742806871 | 4.13E–11 | –1.346990861 | 2.53E–05 |
| SEZ6L2 | 8.486954783 | –1.397656937 | 1.28E–08 | 2.065753561 | 1.62E–07 | 3.463410497 | 3.14E–35 |
| DHDH | 3.645465178 | –1.422563251 | 1.86E–10 | 1.924565805 | 6.56E–10 | 3.347129056 | 6.40E–33 |
| RPSAP52 | 3.843285951 | –1.425249557 | 6.82E–13 | 3.035741512 | 2.13E–17 | 4.46099107 | 9.72E–71 |
| PCSK1N | 7.108530795 | –1.426337445 | 1.27E–05 | 2.931204194 | 2.29E–08 | 4.357541639 | 2.31E–31 |
| C6orf174 | 7.841118367 | –1.434317531 | 8.13E–12 | 1.312216929 | 6.66E–05 | 2.746534459 | 2.09E–31 |
| HPCAL4 | 7.107764569 | –1.439936137 | 3.35E–07 | 3.047122266 | 4.89E–14 | 4.487058404 | 7.70E–38 |
| MMP1 | 3.072302031 | –1.44234842 | 1.82E–08 | 1.547079052 | 3.39E–05 | 2.989427472 | 1.85E–22 |
| MFAP4 | 7.062677167 | –1.442809228 | 3.89E–08 | –4.41365813 | 1.18E–24 | –2.970848902 | 3.27E–22 |
| SSX1 | 5.406962065 | –1.456265836 | 5.53E–09 | 1.525310101 | 0.000281107 | 2.981575937 | 2.14E–24 |
| BICC1 | 3.986170109 | –1.463093037 | 5.72E–10 | –2.855346837 | 1.44E–15 | –1.3922538 | 7.23E–07 |
| TNFRSF12A | 11.28366917 | –1.475952455 | 4.33E–21 | 1.125925211 | 7.23E–05 | 2.601877666 | 8.69E–38 |
| KIAA1755 | 3.927930829 | –1.485363102 | 2.90E–11 | –2.872580611 | 2.75E–17 | –1.387217508 | 4.30E–08 |
| IQGAP3 | 5.366286364 | –1.487991742 | 1.11E–20 | 1.242958702 | 2.03E–06 | 2.730950444 | 5.29E–39 |
| C3orf50 | 2.721838797 | –1.494046089 | 3.57E–12 | 1.80160834 | 1.48E–08 | 3.295654429 | 4.70E–38 |
| OMD | 3.83628455 | –1.494528145 | 6.31E–06 | –4.932157065 | 1.02E–26 | –3.43762892 | 6.83E–18 |
| C6orf168 | 7.834659081 | –1.497712261 | 8.85E–23 | 1.683704188 | 1.47E–13 | 3.18141645 | 4.48E–61 |
| SFTPA2 | 3.581154116 | –1.521985438 | 6.81E–05 | 1.803204172 | 6.52E–06 | 3.32518961 | 7.23E–13 |
| ALDH3B1 | 9.190855645 | –1.53751215 | 7.10E–24 | 1.011722475 | 3.02E–06 | 2.549234626 | 1.04E–44 |
| SLC30A3 | 4.554644551 | –1.544239276 | 5.66E–06 | 1.009436448 | 0.022276581 | 2.553675724 | 8.89E–11 |
| L1CAM | 4.105765596 | –1.549496574 | 2.99E–10 | –2.665607037 | 9.56E–15 | –1.116110463 | 7.58E–05 |
| SSTR3 | 2.617605109 | –1.558963794 | 2.99E–12 | 1.062532698 | 0.000597215 | 2.621496492 | 4.70E–23 |
| IL7R | 4.08720034 | –1.562392878 | 1.14E–08 | –3.234551803 | 5.87E–15 | –1.672158925 | 5.93E–07 |
| NPW | 3.765276637 | –1.567670941 | 7.22E–08 | 1.906225475 | 1.31E–06 | 3.473896416 | 2.23E–23 |
| CLDN9 | 7.281804098 | –1.571351595 | 7.15E–12 | 1.646672219 | 0.000138189 | 3.218023813 | 8.97E–36 |
| MFAP5 | 4.37663423 | –1.576225635 | 2.94E–06 | –4.312229843 | 1.81E–20 | –2.736004208 | 8.49E–12 |
| ABCC3 | 10.3657708 | –1.585928841 | 2.70E–17 | 1.215970769 | 0.00052715 | 2.801899609 | 6.10E–45 |
| REN | 3.853778356 | –1.586075935 | 6.14E–10 | 2.856046415 | 3.20E–15 | 4.44212235 | 6.45E–44 |
| QPCT | 10.08996234 | –1.605279208 | 1.83E–16 | 1.755207231 | 1.01E–07 | 3.360486438 | 1.08E–50 |
| AIM1L | 3.412449737 | –1.609760935 | 6.78E–17 | 1.013946929 | 9.75E–06 | 2.623707864 | 1.43E–26 |
| C1QL1 | 5.201042789 | –1.610213058 | 9.80E–11 | 2.004156249 | 6.67E–09 | 3.614369307 | 5.87E–32 |
| HCN4 | 5.787517713 | –1.611275961 | 3.65E–11 | 3.212386326 | 3.50E–15 | 4.823662287 | 3.32E–54 |
| UBD | 5.208143949 | –1.612660673 | 5.56E–06 | –2.793592901 | 1.05E–06 | –1.180932228 | 0.008215817 |
| SOX11 | 3.293931847 | –1.625465784 | 2.87E–09 | 1.499816443 | 7.17E–05 | 3.125282227 | 2.23E–21 |
| CDH15 | 2.363645641 | –1.62851333 | 1.48E–12 | 1.244042847 | 4.76E–05 | 2.872556177 | 4.95E–25 |
| MRC2 | 11.51299146 | –1.639137173 | 1.46E–16 | 1.271615153 | 4.63E–05 | 2.910752326 | 1.23E–38 |
| RASD2 | 8.331804089 | –1.646912271 | 2.03E–24 | 1.464234653 | 1.06E–08 | 3.111146925 | 7.62E–57 |
| CAPN6 | 3.82365956 | –1.648608995 | 3.80E–08 | –4.71269072 | 1.13E–25 | –3.064081726 | 9.37E–18 |
| BAIAP3 | 8.009200791 | –1.652827092 | 3.94E–14 | 1.863968858 | 7.91E–09 | 3.516795949 | 5.60E–40 |
| C1orf187 | 2.882573689 | –1.65392229 | 1.81E–16 | 1.535933403 | 1.34E–08 | 3.189855693 | 3.06E–38 |
| SERPINA3 | 4.02890599 | –1.667640601 | 1.78E–07 | –3.779654823 | 4.64E–15 | –2.112014223 | 9.97E–09 |
| DBC1 | 6.244410653 | –1.68872723 | 5.31E–11 | 1.241864249 | 0.000498939 | 2.930591478 | 2.00E–22 |
| FAIM2 | 3.079910586 | –1.690939847 | 8.46E–12 | –3.070173321 | 5.95E–18 | –1.379233474 | 1.31E–06 |
| C1S | 9.424758781 | –1.694103139 | 2.83E–12 | –3.589365091 | 1.51E–22 | –1.895261952 | 7.29E–12 |
| WISP2 | 3.608122122 | –1.69763062 | 2.07E–08 | –3.832847485 | 3.05E–19 | –2.135216866 | 2.51E–09 |
| RUNX1 | 9.662948546 | –1.699016409 | 3.25E–29 | 1.026209958 | 2.97E–06 | 2.725226367 | 2.84E–44 |
| SFRP4 | 6.991982984 | –1.707760388 | 1.92E–07 | –3.298429685 | 2.06E–15 | –1.590669297 | 5.08E–05 |
| APOD | 7.353149952 | –1.710317804 | 1.63E–13 | –4.54317184 | 5.38E–32 | –2.832854036 | 1.66E–24 |
| RYR1 | 8.953959279 | –1.713360804 | 6.33E–19 | 1.334502549 | 1.06E–05 | 3.047863354 | 5.95E–43 |
| PDGFRA | 6.476424754 | –1.718104037 | 7.37E–12 | –3.650602893 | 8.60E–22 | –1.932498856 | 8.30E–12 |
| PCDH9 | 4.670654931 | –1.727978157 | 2.86E–17 | –3.033001272 | 9.50E–24 | –1.305023115 | 6.29E–08 |
| CTXN1 | 8.009058961 | –1.763385999 | 3.63E–23 | 1.457296775 | 2.65E–08 | 3.220682774 | 1.19E–49 |
| PLCD3 | 10.84364841 | –1.772353965 | 1.27E–23 | 1.495115453 | 3.03E–09 | 3.267469418 | 9.02E–53 |
| PLEKHN1 | 6.626971012 | –1.773726614 | 4.33E–21 | 2.631863868 | 2.24E–16 | 4.405590483 | 8.19E–69 |
| IGSF1 | 11.71630903 | –1.78107502 | 6.85E–08 | 2.052720867 | 4.16E–05 | 3.833795887 | 1.23E–25 |
| TMEM130 | 5.742181813 | –1.812281079 | 1.31E–09 | 1.143840169 | 0.006830506 | 2.956121248 | 1.45E–17 |
| CD164L2 | 5.193205691 | –1.817896375 | 1.50E–12 | 1.913240322 | 1.16E–06 | 3.731136696 | 3.26E–34 |
| COLEC12 | 6.317141885 | –1.829362368 | 1.91E–19 | –3.496562939 | 6.22E–32 | –1.667200571 | 3.00E–12 |
| SLC7A4 | 3.148586642 | –1.840295097 | 2.88E–17 | 1.021679263 | 0.001690122 | 2.86197436 | 8.14E–30 |
| PDE4C | 7.543782974 | –1.845592352 | 1.27E–17 | 1.851950899 | 4.25E–07 | 3.697543252 | 5.27E–49 |
| ZCCHC16 | 6.129194246 | –1.849487461 | 2.22E–07 | 3.490855022 | 2.97E–10 | 5.340342483 | 1.57E–36 |
| PROS1 | 11.94787525 | –1.856710215 | 1.07E–24 | 1.395254207 | 2.46E–10 | 3.251964422 | 3.27E–47 |
| KCNJ2 | 8.629050602 | –1.865501275 | 3.55E–20 | 1.608687068 | 6.52E–09 | 3.474188343 | 4.60E–45 |
| ASPHD1 | 4.492365251 | –1.868434973 | 2.29E–14 | 2.197765042 | 5.95E–10 | 4.066200015 | 5.47E–40 |
| CAMK2N1 | 10.89390796 | –1.870821948 | 1.55E–19 | 1.268003517 | 1.07E–05 | 3.138825466 | 3.03E–38 |
| DUSP13 | 2.69928433 | –1.872481828 | 9.30E–14 | 1.416786077 | 6.86E–07 | 3.289267905 | 1.04E–25 |
| C10orf55 | 4.74734646 | –1.881624415 | 9.28E–20 | 1.084680643 | 4.84E–05 | 2.966305058 | 4.96E–32 |
| CLDN1 | 12.44363484 | –1.886245621 | 8.20E–16 | 1.732571103 | 3.30E–05 | 3.618816724 | 1.56E–42 |
| PRSS3 | 2.760638938 | –1.902404307 | 2.14E–11 | 1.603076953 | 3.36E–06 | 3.50548126 | 1.60E–24 |
| LPAR5 | 9.663722153 | –1.918037156 | 6.22E–21 | 1.595606392 | 7.02E–06 | 3.513643548 | 7.31E–53 |
| CP | 5.316216757 | –1.932134214 | 3.61E–10 | –3.767453822 | 6.07E–16 | –1.835319607 | 1.14E–07 |
| LEMD1 | 5.617734223 | –1.96252133 | 3.62E–24 | 1.045258786 | 0.000323008 | 3.007780115 | 5.21E–41 |
| XKRX | 6.612691837 | –1.966019785 | 1.34E–16 | 1.750388726 | 2.20E–06 | 3.716408511 | 5.80E–41 |
| PRR15 | 9.790867813 | –1.977623745 | 2.93E–10 | 4.097384446 | 5.97E–15 | 6.075008191 | 1.61E–56 |
| TREM1 | 6.011134814 | –1.989227823 | 2.75E–15 | 1.227990292 | 0.003065686 | 3.217218115 | 1.59E–27 |
| DCN | 9.556657469 | –2.005988726 | 3.78E–12 | –4.918362662 | 5.24E–25 | –2.912373936 | 2.38E–19 |
| TRY6 | 2.468996697 | –2.006384441 | 1.77E–10 | 1.0480905 | 0.001517874 | 3.054474941 | 7.42E–16 |
| RDH5 | 7.71983573 | –2.006606414 | 1.56E–19 | 1.55493853 | 2.70E–06 | 3.561544944 | 1.43E–43 |
| LRRC15 | 4.06796788 | –2.042329423 | 7.60E–08 | –4.817187392 | 2.77E–25 | –2.77485797 | 1.49E–09 |
| CDH3 | 10.61382771 | –2.061524377 | 2.37E–22 | 2.12513314 | 2.56E–08 | 4.186657517 | 2.29E–67 |
| DOK7 | 6.841576162 | –2.092841103 | 1.09E–19 | 1.101920504 | 0.00189577 | 3.194761607 | 8.29E–32 |
| SPTBN2 | 7.785780872 | –2.118222136 | 6.53E–26 | 1.764056236 | 4.06E–09 | 3.882278372 | 5.80E–55 |
| IL1F5 | 3.669556547 | –2.148068318 | 2.34E–20 | 1.547774378 | 8.33E–07 | 3.695842696 | 4.72E–38 |
| RXRG | 9.663387804 | –2.151748123 | 3.60E–11 | 3.774612604 | 3.40E–10 | 5.926360726 | 6.30E–59 |
| FIBCD1 | 4.772968167 | –2.16369903 | 1.58E–12 | 3.113906493 | 3.82E–12 | 5.277605524 | 1.05E–42 |
| LDLR | 8.52211624 | –2.185181279 | 2.17E–33 | –1.175090812 | 7.61E–07 | 1.010090467 | 1.82E–06 |
| PRMT8 | 3.966210495 | –2.185713249 | 5.71E–08 | 1.130933717 | 0.012369183 | 3.316646966 | 5.52E–12 |
| SCEL | 9.551222529 | –2.230354546 | 4.40E–20 | 1.990053842 | 9.60E–09 | 4.220408387 | 1.34E–45 |
| EPHB3 | 8.086567569 | –2.260434535 | 8.30E–32 | 1.31919037 | 1.65E–06 | 3.579624905 | 1.78E–53 |
| PRSS1 | 3.840709232 | –2.269595304 | 1.70E–09 | 2.226272925 | 7.92E–07 | 4.495868228 | 3.18E–23 |
| FAM178B | 4.345314871 | –2.278206519 | 1.12E–17 | 2.639528746 | 1.81E–10 | 4.917735265 | 7.71E–53 |
| ZCCHC12 | 11.66056967 | –2.284612976 | 5.07E–09 | 4.477319537 | 1.90E–11 | 6.761932513 | 6.63E–49 |
| C3 | 12.15527331 | –2.287396246 | 9.35E–18 | –1.171972801 | 0.001001078 | 1.115423445 | 0.000248042 |
| HMGA2 | 8.752976637 | –2.29831632 | 1.64E–13 | 3.958495233 | 8.47E–12 | 6.256811553 | 1.19E–65 |
| HSD11B1 | 3.29533143 | –2.299138944 | 3.25E–20 | –3.485502872 | 3.85E–21 | –1.186363928 | 2.33E–05 |
| CPNE4 | 4.478595265 | –2.306554 | 3.89E–10 | 1.662592746 | 0.001002792 | 3.969146747 | 1.14E–19 |
| CCL19 | 4.13083191 | –2.318954438 | 5.56E–08 | –4.614307273 | 6.69E–13 | –2.295352835 | 1.48E–05 |
| DPP4 | 10.66335627 | –2.319579942 | 1.63E–17 | 2.275937089 | 1.53E–07 | 4.595517032 | 7.85E–47 |
| LCN6 | 4.845901275 | –2.322677193 | 2.89E–09 | 1.683563325 | 0.000145489 | 4.006240518 | 3.65E–17 |
| EPHA10 | 6.562387291 | –2.368477143 | 2.29E–22 | 1.741314359 | 1.53E–07 | 4.109791502 | 6.89E–44 |
| SIGLEC15 | 3.708474273 | –2.381161232 | 1.86E–17 | 1.149207645 | 0.005384442 | 3.530368877 | 1.76E–27 |
| TRPC5 | 6.183370078 | –2.395944989 | 4.65E–09 | 3.532163742 | 1.42E–09 | 5.928108731 | 4.30E–33 |
| GRHL3 | 5.816801554 | –2.541275605 | 9.38E–25 | 3.213956972 | 5.67E–17 | 5.755232577 | 6.00E–71 |
| KCNQ3 | 5.734309467 | –2.554545971 | 3.92E–29 | 2.051132877 | 1.15E–12 | 4.605678848 | 4.06E–55 |
| MYBPH | 3.148521931 | –2.590335073 | 5.57E–18 | 1.044026877 | 0.001203398 | 3.634361949 | 1.17E–22 |
| C7 | 7.673434797 | –2.656347395 | 3.13E–13 | –5.325875815 | 1.35E–19 | –2.66952842 | 1.58E–11 |
| BIRC7 | 4.485020456 | –2.669752292 | 9.74E–15 | 1.868054497 | 5.32E–05 | 4.537806788 | 3.39E–28 |
| FUT3 | 3.409507732 | –2.67193596 | 1.29E–21 | 1.44218836 | 1.16E–06 | 4.11412432 | 6.24E–32 |
| TMEM163 | 7.565014885 | –2.704339847 | 1.97E–24 | 1.116343775 | 0.000648669 | 3.820683622 | 2.61E–31 |
| IGFN1 | 6.787724503 | –2.705287433 | 3.78E–09 | –4.761319042 | 4.98E–14 | –2.056031608 | 0.000101384 |
| LCN2 | 7.828926052 | –2.710891759 | 1.95E–16 | 1.508015232 | 0.000415624 | 4.218906991 | 5.56E–26 |
| CHIT1 | 7.445373302 | –2.715936297 | 6.08E–15 | –1.379937149 | 0.008500852 | 1.335999148 | 0.001000091 |
| PSG1 | 3.499568112 | –2.761901016 | 3.09E–16 | 1.528287949 | 3.84E–05 | 4.290188965 | 4.15E–25 |
| ICAM5 | 7.422968004 | –2.773997645 | 2.81E–23 | 1.527207778 | 0.001120156 | 4.301205423 | 8.85E–44 |
| MMP12 | 2.794033253 | –2.800439197 | 1.25E–19 | –1.226186463 | 0.002759925 | 1.574252733 | 4.89E–05 |
| PVRL4 | 8.256023458 | –2.810599953 | 1.12E–28 | 1.968636411 | 1.27E–07 | 4.779236364 | 1.16E–54 |
| ADAMTS14 | 5.059171574 | –2.84492367 | 3.14E–27 | 1.01224032 | 0.001887892 | 3.85716399 | 4.32E–31 |
| GRM4 | 5.336364809 | –2.864309793 | 2.23E–16 | 2.311357066 | 2.51E–06 | 5.175666859 | 1.33E–35 |
| CSF2 | 3.096337603 | –2.884248914 | 4.12E–30 | 1.001995199 | 0.000429609 | 3.886244113 | 4.50E–38 |
| GJB3 | 7.46922135 | –2.896067796 | 3.26E–27 | 1.978991131 | 4.66E–06 | 4.875058927 | 4.45E–55 |
| ARSI | 2.905034147 | –2.898266649 | 1.93E–26 | –1.150151967 | 3.44E–05 | 1.748114682 | 6.76E–08 |
| CST2 | 3.060575286 | –2.935586584 | 7.49E–17 | 1.099499149 | 0.00154906 | 4.035085734 | 1.50E–20 |
| CFB | 10.07224149 | –2.951725784 | 3.06E–24 | –1.469820993 | 0.000784975 | 1.481904791 | 2.09E–06 |
| CCL22 | 4.938310502 | –2.958006693 | 5.18E–25 | –1.728477417 | 1.90E–05 | 1.229529277 | 0.000307643 |
| LIPH | 8.485931656 | –2.995825873 | 1.35E–22 | 3.962891385 | 5.21E–12 | 6.958717258 | 1.18E–77 |
| NMU | 5.349207193 | –3.022248534 | 6.13E–14 | 2.106809569 | 2.70E–05 | 5.129058103 | 6.03E–25 |
| CLDN16 | 7.776262359 | –3.023704377 | 2.69E–18 | 2.145013865 | 1.24E–05 | 5.168718242 | 4.27E–36 |
| CATSPER1 | 3.897340433 | –3.064740416 | 1.16E–26 | 1.144065389 | 0.00176363 | 4.208805805 | 5.09E–35 |
| ACTBL2 | 3.160309268 | –3.071239217 | 6.36E–33 | 1.042633471 | 0.00014476 | 4.113872688 | 5.05E–39 |
| DMBX1 | 3.145088597 | –3.093923693 | 1.05E–18 | 1.135558708 | 0.000148333 | 4.229482401 | 3.16E–22 |
| SLC6A20 | 4.798247522 | –3.172385244 | 5.13E–17 | 1.462617334 | 0.001110525 | 4.635002578 | 2.50E–24 |
| PPP1R1B | 6.345161106 | –3.173819555 | 3.11E–22 | 2.174238178 | 7.74E–06 | 5.348057733 | 4.52E–44 |
| LAMB3 | 10.61279751 | –3.185092156 | 5.18E–30 | 1.155974145 | 0.00454641 | 4.341066301 | 2.54E–42 |
| GABRB2 | 9.66515018 | –3.193052409 | 3.79E–30 | 4.287074604 | 4.50E–24 | 7.480127012 | 2.69E–83 |
| S100B | 5.227085376 | –3.200053334 | 6.13E–23 | –1.850044112 | 4.97E–06 | 1.350009222 | 0.000245816 |
| NGEF | 8.503253119 | –3.219738834 | 3.11E–28 | 2.2166878 | 9.48E–07 | 5.436426634 | 5.54E–57 |
| STRA6 | 7.536565326 | –3.274609991 | 4.29E–23 | 2.389999198 | 2.29E–06 | 5.664609189 | 3.93E–47 |
| ARHGAP36 | 8.063784001 | –3.329685605 | 7.61E–10 | 4.372591202 | 6.54E–08 | 7.702276807 | 1.64E–34 |
| RASGRF1 | 6.35556206 | –3.340251478 | 1.00E–23 | 1.560042684 | 0.000889007 | 4.900294162 | 4.22E–35 |
| SERPINA1 | 13.97208088 | –3.347721751 | 4.69E–33 | 1.045563118 | 0.008141256 | 4.393284869 | 1.76E–44 |
| SFRP2 | 7.5685554 | –3.358137508 | 2.38E–15 | –4.992447199 | 1.39E–17 | –1.63430969 | 0.000896524 |
| KRT15 | 5.503566928 | –3.380639979 | 1.09E–29 | 2.07529649 | 1.67E–06 | 5.45593647 | 1.57E–52 |
| ESPN | 5.340893034 | –3.455938561 | 3.52E–24 | 2.053384312 | 2.51E–05 | 5.509322873 | 2.78E–43 |
| KLK11 | 5.28315049 | –3.508030452 | 9.76E–13 | 1.798789279 | 0.003175094 | 5.306819731 | 1.34E–19 |
| ADCY8 | 4.614735007 | –3.533430934 | 6.62E–19 | 2.023233142 | 2.33E–05 | 5.556664075 | 7.78E–31 |
| KLK6 | 3.747977098 | –3.563812068 | 9.86E–21 | 1.229696825 | 0.001232396 | 4.793508893 | 9.36E–25 |
| LOC400794 | 5.540774446 | –3.572123883 | 8.01E–25 | 2.696458943 | 2.11E–08 | 6.268582827 | 2.28E–51 |
| CCL18 | 7.317167047 | –3.573512233 | 9.86E–28 | –1.672339177 | 0.001585203 | 1.901173056 | 1.01E–07 |
| FN1 | 16.5189005 | –3.791945965 | 1.37E–34 | 1.387573036 | 4.61E–05 | 5.179519001 | 2.14E–44 |
| IL1RL1 | 6.305090441 | –3.878985653 | 7.48E–25 | –2.384806625 | 3.49E–07 | 1.494179029 | 0.001612935 |
| SYTL5 | 7.469621074 | –3.914045782 | 3.55E–30 | 2.232554173 | 1.36E–05 | 6.146599955 | 2.58E–53 |
| CXCL14 | 9.639685447 | –3.952231881 | 2.21E–18 | –1.93621346 | 0.001347999 | 2.016018421 | 5.78E–05 |
| MUC21 | 6.066486624 | –4.028635997 | 1.98E–23 | 2.402439566 | 8.77E–06 | 6.431075563 | 2.35E–39 |
| TMEM92 | 4.485349996 | –4.126972544 | 1.94E–32 | 1.416689638 | 0.000346983 | 5.543662182 | 2.46E–40 |
| KLK10 | 8.67098633 | –4.195424379 | 2.56E–22 | 1.579820838 | 0.011519768 | 5.775245217 | 2.40E–32 |
| B3GNT3 | 7.513903367 | –4.698635959 | 1.21E–31 | 1.063281816 | 0.042498779 | 5.761917776 | 1.66E–34 |
| TMPRSS4 | 8.102882755 | –4.736650488 | 9.48E–34 | 1.76822058 | 0.000347084 | 6.504871068 | 5.52E–44 |
| SLC27A6 | 8.306267997 | –4.982911793 | 2.38E–33 | –1.456437976 | 0.017905016 | 3.526473818 | 2.63E–16 |
| SYT12 | 8.489871446 | –5.09305182 | 2.41E–33 | 2.206952182 | 4.34E–05 | 7.300004002 | 3.75E–47 |
| SFTPB | 11.52642597 | –5.312188634 | 9.48E–34 | –1.125548306 | 0.043156747 | 4.186640327 | 1.20E–17 |
| TMPRSS6 | 7.444079358 | –5.470904727 | 1.07E–33 | 1.657424494 | 0.002245598 | 7.128329221 | 7.51E–41 |
FC, fold changes; FVPTC, follicular variant papillary thyroid carcinoma; DEG, differentially expressed gene; NT, normal tissues; cPTC, conventional papillary thyroid carcinoma; RSEM, RNAseq by expectation-maximization.
Acknowledgments
Funding: This work was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.38). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg 2014;140:317-22. [Crossref] [PubMed]
- Shaha AR, Tuttle RM. Thyroid cancer staging and genomics. Ann Transl Med 2019;7:S49. [Crossref] [PubMed]
- Li M, Zhao B, Qu W, et al. Uncovering the potential miRNAs and mRNAs in follicular variant of papillary thyroid carcinoma in the Cancer Genome Atlas database. Transl Cancer Res 2019;8:1158-69. [Crossref]
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11. Lyon: International Agency for Research on Cancer, 2013.
- SEER. Cancer stat facts: thyroid cancer. Available online: http://seer.cancer.gov/statfacts/html/thyro.html
- Vuong HG, Kondo T, Duong UNP, et al. Prognostic impact of vascular invasion in differentiated thyroid carcinoma: a systematic review and meta-analysis. Eur J Endocrinol 2017;177:207-16. [Crossref] [PubMed]
- Lloyd RV, Buehler D, Khanafshar E. Papillary thyroid carcinoma variants. Head Neck Pathol 2011;5:51-6. [Crossref] [PubMed]
- Xu B, Ghossein R. Evolution of the histologic classification of thyroid neoplasms and its impact on clinical management. Eur J Surg Oncol 2018;44:338-47. [Crossref] [PubMed]
- Tunca F, Sormaz IC, Iscan Y, et al. Comparison of histopathological features and prognosis of classical and follicular variant papillary thyroid carcinoma. J Endocrinol Invest 2015;38:1327-34. [Crossref] [PubMed]
- Shi X, Liu R, Basolo F, et al. Differential clinicopathological risk and prognosis of major papillary thyroid cancer variants. J Clin Endocrinol Metab 2016;101:264-74. [Crossref] [PubMed]
- Kim SK, Park I, Woo JW, et al. Follicular and diffuse sclerosing variant papillary thyroid carcinomas as independent predictive factors of loco-regional recurrence: a comparison study using propensity score matching. Thyroid 2016;26:1077-84. [Crossref] [PubMed]
- Tallini G, Tuttle RM, Ghossein RA. The history of the follicular variant of papillary thyroid carcinoma. J Clin Endocrinol Metab 2017;102:15-22. [PubMed]
- Nikiforov YE, Seethala RR, Tallini G, et al. Nomenclature revision for encapsulated follicular variant of papillary thyroid carcinoma: a paradigm shift to reduce overtreatment of indolent tumors. JAMA Oncol 2016;2:1023-9. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676-90. [Crossref] [PubMed]
- Li H, Liu JW, Liu S, et al. Bioinformatics-based identification of methylated-differentially expressed genes and related pathways in gastric cancer. Dig Dis Sci 2017;62:3029-39. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Li J, Fine JP. ROC analysis with multiple classes and multiple tests: methodology and its application in microarray studies. Biostatistics 2008;9:566-76. [Crossref] [PubMed]
- Stokowy T, Gawel D, Wojtas B. Differences in miRNA and mRNA profile of papillary thyroid cancer variants. Int J Endocrinol 2016;2016:1427042.
- Saiselet M, Gacquer D, Spinette A, et al. New global analysis of the microRNA transcriptome of primary tumors and lymph node metastases of papillary thyroid cancer. BMC Genomics 2015;16:828. [Crossref] [PubMed]
- McFadden DG, Dias-Santagata D, Sadow PM, et al. Identification of oncogenic mutations and gene fusions in the follicular variant of papillary thyroid carcinoma. J Clin Endocrinol Metab 2014;99:E2457-62. [Crossref] [PubMed]
- Kim TH, Lee M, Kwon AY, et al. Molecular genotyping of the non-invasive encapsulated follicular variant of papillary thyroid carcinoma. Histopathology 2018;72:648-61. [Crossref] [PubMed]
- Zhao L, Dias-Santagata D, Sadow PM, et al. Cytological, molecular, and clinical features of noninvasive follicular thyroid neoplasm with papillary-like nuclear features versus invasive forms of follicular variant of papillary thyroid carcinoma. Cancer Cytopathol 2017;125:323-31. [Crossref] [PubMed]
- Yoo SK, Lee S, Kim SJ, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet 2016;12:e1006239. [Crossref] [PubMed]
- Marlow R, Strickland P, Lee JS, et al. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res 2008;68:7819-27. [Crossref] [PubMed]
- Dickinson RE, Dallol A, Bieche I, et al. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer 2004;91:2071-8. [Crossref] [PubMed]
- Kim M, Kim JH, Baek SJ, et al. Specific expression and methylation of SLIT1, SLIT2, SLIT3, and miR-218 in gastric cancer subtypes. Int J Oncol 2016;48:2497-507. [Crossref] [PubMed]
- Amodeo V, Deli A, Betts J, et al. A PML/slit axis controls physiological cell migration and cancer invasion in the CNS. Cell Rep 2017;20:411-26. [Crossref] [PubMed]
- Zhang L, Wang D, Li Y, et al. CCL21/CCR7 axis contributed to CD133+ pancreatic cancer stem-like cell metastasis via EMT and Erk/NF-κB pathway. PLoS One 2016;11:e0158529. [Crossref] [PubMed]
- Xiong Y, Huang F, Li X, et al. CCL21/CCR7 interaction promotes cellular migration and invasion via modulation of the MEK/ERK1/2 signaling pathway and correlates with lymphatic metastatic spread and poor prognosis in urinary bladder cancer. Int J Oncol 2017;51:75-90. [Crossref] [PubMed]
- Lu LL, Chen XH, Zhang G, et al. CCL21 facilitates chemoresistance and cancer stem cell-like properties of colorectal cancer cells through AKT/GSK-3β/snail signals. Oxid Med Cell Longev 2016;2016:5874127.
- Zhang YY, Liu ZB, Ye XG, et al. Iodine regulates G2/M progression induced by CCL21/CCR7 interaction in primary cultures of papillary thyroid cancer cells with RET/PTC expression. Mol Med Rep 2016;14:3941-6. [Crossref] [PubMed]
- Sand LG, Berghuis D, Szuhai K, et al. Expression of CCL21 in Ewing sarcoma shows an inverse correlation with metastases and is a candidate target for immunotherapy. Cancer Immunol Immunother 2016;65:995-1002. [Crossref] [PubMed]
- Ivanova AV, Goparaju CM, Ivanov SV, et al. Protumorigenic role of HAPLN1 and its IgV domain in malignant pleural mesothelioma. Clin Cancer Res 2009;15:2602-11. [Crossref] [PubMed]
- Mebarki S, Désert R, Sulpice L, et al. De novo HAPLN1 expression hallmarks Wnt-induced stem cell and fibrogenic networks leading to aggressive human hepatocellular carcinomas. Oncotarget 2016;7:39026-43. [Crossref] [PubMed]
- Zhuang Z, Jian P, Longjiang L, et al. Oral cancer cells with different potential of lymphatic metastasis displayed distinct biologic behaviors and gene expression profiles. J Oral Pathol Med 2010;39:168-75. [Crossref] [PubMed]
- Wang O, Zheng Z, Wang Q, et al. ZCCHC12, a novel oncogene in papillary thyroid cancer. J Cancer Res Clin Oncol 2017;143:1679-86. [Crossref] [PubMed]
- Santarpia L, Myers JN, Sherman SI, et al. Genetic alterations in the RAS/RAF/mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways in the follicular variant of papillary thyroid carcinoma. Cancer 2010;116:2974-83. [Crossref] [PubMed]
- Eszlinger M, Lau L, Ghaznavi S, et al. Molecular profiling of thyroid nodule fine-needle aspiration cytology. Nat Rev Endocrinol 2017;13:415-24. [Crossref] [PubMed]
- Liu YY, Morreau H, Kievit J, et al. Combined immunostaining with galectin-3, fibronectin-1, CITED-1, Hector Battifora mesothelial-1, cytokeratin-19, peroxisome proliferator-activated receptor-{gamma}, and sodium/iodide symporter antibodies for the differential diagnosis of non-medullary thyroid carcinoma. Eur J Endocrinol 2008;158:375-84. [Crossref] [PubMed]
- Martins MB, Marcello MA, Morari EC, et al. Clinical utility of KAP-1 expression in thyroid lesions. Endocr Pathol 2013;24:77-82. [Crossref] [PubMed]
- Hoftijzer HC, Liu YY, Morreau H, et al. Retinoic acid receptor and retinoid X receptor subtype expression for the differential diagnosis of thyroid neoplasms. Eur J Endocrinol 2009;160:631-8. [Crossref] [PubMed]


