
FOXO3/TRIM22 axis abated the antitumor effect of gemcitabine in non-small cell lung cancer via autophagy induction
Introduction
Lung cancer is one of the leading causes of death around the world now. Non-small cell lung cancer (NSCLC) accounts for more than 80% of the total lung cancer (1). Gemcitabine (GEM)-based chemotherapy is the first-line therapeutic approach for the NSCLC treatment (2). However, owing to acquired chemo-resistance, the therapeutic effect is not satisfactory, and the prognosis of NSCLC patients receiving GEM treatment is still poor (3). Based on this, the molecular mechanism underlying chem-resistance requires further investigation to improve the clinical outcomes of NSCLC patients.
The underlying mechanisms of GEM-resistance are not fully understood yet. Some studies have demonstrated that chem-resistance is promoted by activation of autophagy in different cancer cell lines. Furthermore, autophagy inhibition increases chemo-sensitivity to anticancer drugs not only in vitro but also in vivo (4-6). Autophagy often plays a dual role in determining cell death/survival decisions, it can act as an either alternative cell death pathway or a cytoprotective role which depends on different context (7). In recent years, autophagy is largely believed to function as a pro-survival process due to its critical role in cellular energy and nutrition homeostasis, and autophagy inhibition compromises cell viability which has been considered as a potential molecular mechanism of drug resistance in tumors (8). Autophagy occurs at basal levels under physiological conditions and can also be upregulated in response to stressful stimuli such as hypoxia, nutritional deprivation, DNA damage, and cytotoxic agents (9). Autophagy frequently occurs during tumorigenesis and chemotherapy. Generally, constructive autophagy protects tumor cells during chemotherapy, leading to drug resistance and refractory cancer (10). Therefore, elucidation of autophagic signaling mechanisms with regards to specific types of cancer and therapies may be of great significance to develop more effective chemotherapeutic combinations for cancer treatment (11).
Tripartite motif protein 22 (TRIM22) is a member of tripartite motif family protein which involved in innate immunity and autophagy process. TRIM22 serves as an important oncogene and a promoter of cell proliferation and invasion through AKT/GSK3β/β-catenin induced epithelial stromal transformation (EMT) in NSCLC (12). By comparing and analyzing the GEO database (GSEA6914), it was found that the expression of TRIM22 in GEM-resistant (GR) non-small lung cancer Calu3 cells was significantly higher than that of its parent cell line which suggest TRIM22 may play a role in chemo-resistance of NSCLC (13). Studies have shown that TRIM22 can improve autophagy in mycobacterium tuberculosis infected macrophages (14). However, up to now, whether TRIM22 affects GEM resistance in lung adenocarcinoma cells by regulating autophagy has not been studied.
In this study, we found that the expression of TRIM22 was significantly upregulated in GR lung adenocarcinoma cell line A549. To some extent, it was due to the decreased expression of FOXO3, which is a transcription inhibitor of TRIM22. Due to the enhancement of pro-survival autophagy induced by TRIM22, the A549 cells became less sensitive to GEM to a certain degree.
Methods
Cell culture and drugs
The NSCLC GR cell line A549/GR and its parental cell line A549 were purchased from Procell (Wuhan, China) and were cultured under the condition of DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS, GIBCO) and 1% antibiotics (100 U/mL penicillin and 100 µg/mL streptomycin sulfates) at 37 °C in humidified atmosphere of 5% CO2. During the drug treatment, A549 parental cells were treated with 5 µM GEM, the A549/GR cells were treated with 5 µM GEM. GEM, 3-MA and rapamycin were all obtained from Selleck (Shanghai, China).
Clinical patient serum sample collection and ethical statement
The collection process of clinical serum samples involved in this research was approved and executed strictly in accordance with rules and regulations of the ethics committee of the Second Xiangya Hospital of Central South University (Ethical approval ID: EA20190001). We recruited and collected the patient serum of who received postoperative chemotherapy with GEM (2–4 cycles) from October 2017 to June 2018 in Xiangya Second Hospital. The patients with recurrence were defined as resistant samples.
Gene silencing and over-expression lentivirus packaging
In order to construct TRIM22 loss-of-function cell model, three siRNA sequences targeting TRIM22 were designed and synthesized in GenePharma (Shanghai, China), and the one with best interference efficiency in A549 cells was used for subsequent experiments. The sequence information of TRIM22-siRNA was: 5'GCTGAGATGTCACATGGCA3'. To obtain TRIM22 gain-of-function cell model, TRIM22 and FOXO3 overexpressed lentivirus were constructed and packaged in Genomeditech (Shanghai, China). Relative transfection and infection were performed according to manufacturer’s instructions. In rescue experiments, A549/GR cells were infected with FOXO3 overexpressed lentivirus (Lv-FOXO3) firstly then infected with TRIM22 overexpressed lentivirus (Lv-TRIM22) when the cells were stable.
RNA isolation and qRT-PCR
Total RNAs were extracted from different group of cells using the TRIzol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR was performed to detect the relative mRNA levels of TRIM22 and FOXO3 using qRT-PCR. ThermoScript One-Step System (Invitrogen, Carlsbad, CA, USA) in the LC480 Real-Time PCR Detection System (Roche) according to the manufacturer’s instructions. All tests were carried out with 3 independent experiments. The primer sequences are as follows: TRIM22-F, 5'-GCACGCTCATCTCAGATCTCC-3'; TRIM22-R, 5'-TTTTGGCTTCTTCAATGTCCAG-3'. FOXO3-F: 5'-CGGACAAACGGCTCACTC-3'; FOXO3-R: 5'-GGACCCGCATGAATCGACTAT-3'; β-actin-F: 5'-CCTCACCCTGAAGTACCCCAT-3', β-actin-R, 5'-GCCAGATTTTCTCCATGTCGTC-3'. The quantification analysis for relative mRNA levels of target genes was performed using the relative quantification comparative CT method.
Western blotting
Western blot was performed to detect the target protein expression of interest according to the methods described in the literature (15). The primary antibodies we used were anti-TRIM22 (13744-1-AP, 1:500, Proteintech, China), anti-FOXO3 (10849-1-AP, 1:800, Proteintech, China), anti-p-FOXO3 (ab31109, 1:500, Abcam, USA), anti-Bax (50599-2-lg, 1:2,000, Proteintech, China), anti-BCl-2 (12789-1-AP, 1:500, Proteintech, China), anti-LC3 (18725-1-AP, 1:300, Proteintech, China), anti-p62 (18420-1-AP, 1:500, Proteintech, China). After incubation with HRP-coupled anti- rabbit IgG antibody at 37 °C for 2 hours. Target proteins were visualized using ECL western blotting detection Kit (FulenGen, Guangzhou, China).
Cell apoptosis and cell cycle analysis with flow cytometer
Annexin V-FITC/propidium iodide (PI) staining assay was used to detect apoptosis after GEM treatment. A549 and A549/GR cells were pretreated with 5 and 20 µM GEM for 24 h. Then, the harvested cells were washed with pre-coded PBS twice and resuspended in Annexin V-FITC binding buffer. Annexin V-FITC/PI (BD Pharmingen, USA) was used to stain cells in darkness. Finally, apoptosis cells were assessed by gating PI and Annexin V-positive cells on a fluorescence-activated cell-sorting (FACS) flow cytometer (BD Pharmingen USA). In the other hand, Cell cycle was also detected by flow cytometry. Cells were harvested at 24 hours. after GEM treatment at the amount of about 5×105. BD Cycletest PLUS DNA Reagent Kit was used in accordance with the instruction. All experiments were repeated for three times. Statistical analysis was performed using Flowjo software. Experiments were performed in triplicate.
Identification of autophagy
A549 parental and GR model cells were infected with AAV- mRFP-GFP-LC3 (Hanbio, Wuhan, China), After 24 h incubation, A549 model cells were treated with 5 µM GEM and A549/GR model cells were treated with 20 µM GEM for another 24 h. respectively. Then, the RFP-LC3 punctate structures were observed and captured using a fluorescent inverted microscope (motif).
MTT assay
The drug resistance was measured using 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Different group of model cells were inoculated into 96-well plates (1,000 cells/well), and 0.5 mg/mL MTT solution was added into each well for 4 h. In order to dissolve the crystalline precipitate, the solution was discarded and replaced with 150 µL of DMSO and the absorbance at 450 nm was detected using a microplate reader. The corresponding negative control was set as normalization 100% survival.
Bi-luciferase reporter assay
According to the ALGEN-PROMO and JASPAR database, the promoter of TRIM22 has the possibility of transcriptional binding to transcription factor FOXO3 which suggested that the expression of TRIM22 may be regulated by the transcription factor FOXO3. Then the synthetic dual luciferase report carrier pGL3.0-TRIM22-promoter and the FOXO3 expression vector pcDNA3.1-FOXO3 were used in luciferase reporter assay (16). During the experiments, the dual luciferase report carrier and pcDNA3.1-FOXO3 vector were co-transfected into 293T-cells. Cells were divided into three groups: the PGL3.0-Basic group, the pGL3.0-TRIM22-promoter group (TRIM22-promoter), and the pGL3.0-TRIM22-promoter and pcDNA3.1-FOXO3 sublet (TRIM22-promoter FOXO3). After 48 hours, cells were collected, and fluorescence values were detected with the Dual Luciferase Reporter Gene Assay Kit (Abnova, China) and the Biotek multifunctional enzyme marker instrument.
Statistical analysis
All experiments were independently repeated three times. Statistical analysis was conducted and presented with GraphPad prism 7.0 software. Differences between two independent groups were evaluated by Student’s t-tests and differences for multiple comparisons were evaluate by one-way ANOVA. P≤0.05 was regarded as statistically significant. Data are presented as means ± SD.
Results
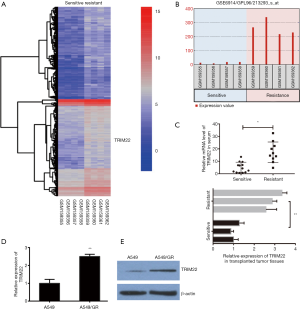
TRIM22 was significantly upregulated in GR non-small lung cancer cells and transplanted tumor tissues of nude mice
In order to search for key genes that may be associated with GEM resistance in NCLCS, we analyzed the GEO data set firstly (GSE 6914). By analyzing of deregulated mRNAs between GR non-small lung cancer Calu3 cells and parental cells, we obtained a series of potential mRNAs with significant differences between the two groups (Log FC >1.0 or <−1.0, P<0.01). It was found that TRIM22 was significantly upregulated in GR Calu3 cells (R) comparing with parental Calu3 cells (S) (Log FC =3.978, adj.P value =0.00068, Figure 1A,B). To further confirm the results of GSE data analysis, on one hand, the relative TRIM22 mRNA levels were detected in clinically collected patient serum samples (sensitive, n=13; resistant, n=10). On the other hand, our group has previously established nude mouse models of transplanted tumors of A549GR and its parental A549 inoculation, the expression of TRIM22 was detected again in transplanted tumor tissues of three of which were considered as GR (inoculated with A549/GR cells) and of three of which were identified as GEM-sensitive (GS) (inoculated with A549 parental cells) respectively. The qRT-PCR showed that the TRIM22 expression in both resistant patient serums and inoculated resistant tissues were obviously stronger than that in sensitive patient serums and inoculated sensitive tissues (Figure 1C). In addition, A549/GR cells and A549 parental cells were also used to detect the TRIM22 expression at the cellular level and a consistent conclusion was reached that TRIM22 was augmented in A549/GR compared with A549 parental cells (Figure 1D,E). These results suggested that TRIM22 may play an important role in GEM sensitivity reduction of NCLCS.
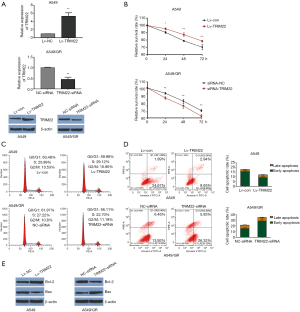
TRIM22 overexpression could attenuate the sensitivity of A549 cells to GEM and its depletion could promote the sensitivity of A549/GR cells to GEM
In order to study the functional role of TRIM22 in GEM-resistance of NCLCS, the gain-of-function cell models of A549 was established via infecting the A549 cells with TRIM22 overexpression lentivirus (Lv-TRIM22), while the loss-of-function cell models was established via TRIM22 silence in A549/GR cells. QPCR and western blot were performed to verify that the TRIM22 functional cell models were obtained successfully (Figure 2A). MTT assay was used to evaluate the cell viability after GEM treatment. The results showed that when the cells were treated with GEM, the TRIM22 over-expressed A549 cells (Lv-TRIM22) exhibited remarkable stronger viability when compared with cells infected with negative control lentivirus (Lv-Con). Meanwhile, the TRIM22 silenced A549/GR cells exhibited significant lower viability when compared with cells transfected with siRNA-NC (Figure 2B) (17). Besides, the cell cycle detection showed that TRIM22 over-expression reduced G0/G1 phase cell cycle arrest slightly in GEM treated A549 and induced G0/G1 phase cell cycle arrest slightly in GEM treated A549/GR respectively (Figure 2C). Then, flow cytometry with annexin V FITC/PI double staining in detection of the GEM induced apoptosis of A549 and A549/GR cells. It was showed that when treated with GEM, TRIM22 over-expression reduced the GEM induced A549 apoptosis obviously compared with cells infected with negative control lentivirus (Lv-Con) and TRIM22 silence increased the GEM induced A549/GR apoptosis significantly compared with A549/GR transfected with siRNA-NC (Figure 2D). Meanwhile, the protein expression of Bax was decreased in TRIM22 over-expressed A549 cells but the BCL-2 was increased conversely. However, in assessing the apoptotic related protein expression in A549/GR cells with TRIM22 knockdown, we got the opposite conclusions (Figure 2E). These results demonstrated that in one hand, TRIM22 could attenuate the sensitivity of A549 cells to GEM. In the other hand, TRIM22 inhibition could promote the sensitivity of A549/GR cells to GEM.
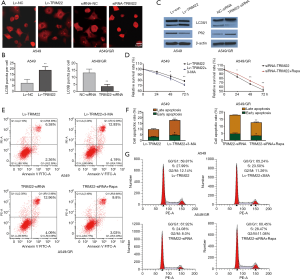
TRIM22 promoted GEM-induced pro-survival autophagy to protected NSCLC cells from apoptotic death
It has been reported that autophagy is involved in GEM-induced A549 lung cell death (17-19). As previously reported, TRIM22 could promote autophagy in macrophage. Based on these, we analyzed whether TRIM22 attenuate the sensitivity of A549 cells to GEM through autophagy regulation. A549 model cells of different groups were transfected with AAv-mRFP-GFP-LC3 virus and later treated with 5 µM GEM for 24 h. The results showed that RFP-LC3 puncta were significantly increased in GEM-treated Lv-TRIM22 cells compared to LV-Con cells. While the RFP-LC3 puncta were significantly decreased in GEM-treated TRIM22 silenced A549/GR cells compared to siRNA-NC transfected A549/GR cells (Figure 3A,B). To confirm this data further, western blot was used to analyze the expression of LC3-II and P62. Protein levels of LC3-II were markedly increased but P62 were obviously decreased by TRIM22 overexpression. Interestingly, knockdown of TRIM22 in A549/GR cells significantly blocks autophagy induction in response to GEM treatment drew reversed outcomes in A549/GR (Figure 3C). In order to further verify TRIM22 promoted GEM-induced pro-survival autophagy to protected NCLCS cells from apoptotic death, we introduced 3-MA (5 mM), an inhibitor of autophagy and rapamycin (100 µM) an autophagy agonist was added to A549 and A549/GR cell cultures respectively for 1 hr. prior to GEM exposure. Cell viability was detected by MTT assay, cell cycle and apoptosis was detected by flow cytometry Results showed that the inhibition of autophagy decreased the cell survival rate but enhanced GEM-induced cell apoptosis in TRIM22 over-expressed A549 cells. While activation of autophagy enhanced the cell survival rate but reduced GEM-induced cell apoptosis in TRIM22 depleted A549/GR cells (Figure 3D,E,F). Although TRIM22 seems to have a slight effect on cell cycle distribution under GEM treatment, the cell cycle distribution was detected again. In TRIM22 over-expressed A549 cells, 3-MA treatment almost abolished the decrease of G0/G1 arrest caused by TRIM22 overexpression and in TRIM22 silenced A549/GR, Rapa addition also abolished the enhancement of G0/G1 arrest caused by TRIM22 knockdown to some extent (Figure 3G). Together with the findings, we drew the conclusion that TRIM22 promoted GEM-induced pro-survival autophagy to protected NCLCS cells from apoptotic death.
TRIM22 expression was negatively transcriptional regulated by FOXO3
We then tried to investigate which transcription factors were involved in TRIM22 upregulation. By utilizing the online programs JASPAR and ALGGEN-PROMO, FOXO3 was found to be a potential transcription factor for TRIM22 gene (Figure 4A). It is also worth mentioning that FOXO3 was also found significantly decreased in GR Calu3 cells comparing with parental Calu3 cells in GSE6917 (Log FC =−2.6172, adj.P value =0.03553, Figure 4B). This suggested that FOXO3 may be a transcriptional suppressor of TRIM22. In addition, A549/GR cells and A549 parental cells were also used to detect the FOXO3 expression at the cellular level and the results demonstrated that FOXO3 was decreased in A549/GR compared with A549 parental cells (Figure 4C). In order to clarify whether FOXO3 regulated the TRIM22 gene expression negatively, A549 and A549/GR cells were transfected with FOXO3 siRNA and infected with FOXO3 overexpression lentivirus respectively. Western blots showed FOXO3 knockdown led to a significant upregulation in the protein expression of TRIM22 with a decrease in the activation form of FOXO3 (p-FOXO3). While, FOXO3 overexpression in A549 cells drew opposite conclusions (Figure 4D). To confirm this interaction between TRIM22-promoter and FOXO3, we performed luciferase reporter assay. The results showed a significant reduction in luciferase levels after co-transfection of pCDNA3.2-FOXO3 with pGL-TRIM22-promoter (Figure 4E). This indicated that FOXO3 could bind with the TRIM22 promoter and inhibit the transcription of TRIM22 to a certain extent.
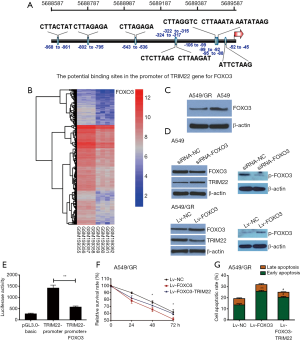
In order to further confirm the regulatory role of FOXO3/TRIM22 axis in GEM susceptibility of A549 cells, rescue experiments were carried out. A549/GR cells were infected with FOXO3 overexpressed lentivirus (Lv-FOXO3) individually or cells were infected with Lv-FOXO3 firstly then infected with TRIM22 overexpressed lentivirus (Lv-TRIM22) when the cells were stable. A549 cells of different groups were treated with GEM and cell viability and apoptosis were estimated again. The results demonstrated that FOXO3 overexpression could decrease the GEM-treated cell survival and increase the cell apoptosis of A549/GR cells. However, if A549/GR was continued to overexpress TRIM22 on the basis of FOXO3 overexpression, the resistance of A549/GR to GEM can be restored to some extent (Figure 4F,G).
Discussion
Autophagy is considered to be a pro-survival mechanism against chemo-sensitivity in these years (20). In the present study, our experiments demonstrated that TRIM22 enhances GEM resistance by regulating cytoprotective autophagy in NSCLC. Targeting FOXO3/TRIM22 axis could enhance lung cancer cell susceptibility to GEM.
Lung cancer is the leading cause of cancer-related mortality which was always diagnosed at the median/advanced stage (21). In view of this situation, the standard therapy is combination chemotherapy (19). GEM which was considered as one of the main anti-cancer drugs is widely used in clinical treatment of lung cancer. However, primary or acquired resistance greatly limits its clinical anti-cancer efficacy. Therefore, it is of great clinical significance to find potential targets and improve the drug sensitivity.
In order to find the key targets for GEM resistance of NSCLC, we compared and analyzed the related GSE database (GSE 6914). Through comparative analysis of mRNA expression profiles of parent cells and GM-resistant cells, TRIM22 was found to be the most significant factor with high data quality. At the same time, we confirmed this trend in transplanted tumor tissues in nude mice and in A549/GR cells using QPCR, so our follow-up study focused on TRIM22.
In our research, reverse modeling was skillfully adopted. TRIM22 gain-of-function cell models was constructed in A549 parental cells and its loss-of-function cell models was constructed in A549/GR cells to clarify the drug-sensitivity modulation of TRIM22. Our results verified that TRIM22 overexpression could attenuate the sensitivity of A549 cells to GEM and its depletion could promote the sensitivity of A549/GR cells to GEM.
In recent years, autophagy is considered to be a survival mechanism to avoid death in responding to a broad range of stress in cancer (22). Although this is still a controversial topic, the importance of autophagy in drug resistance is widely accepted now. In exploring the molecular mechanism of TRIM22 regulating GM sensitivity in lung cancer cells, we associated autophagy. Whether TRIM22 can regulate autophagy is not well documented. Fortunately, our results demonstrated that TRIM22 promoted GEM-induced pro-survival autophagy to protected NSCLC cells from apoptotic death.
It is worth mentioning that, we have a further discovery. Through the comparative analysis of GSE6914 dataset, we found that the classical transcription factor FOXO3 was significantly down-regulated in drug-resistant cells of which was contrary to TRIM22. By utilizing the online programs JASPAR and ALGGEN-PROMO, FOXO3 was found to be a potential transcription factor for TRIM22 gene. This project also confirmed the transcriptional inhibition relationship between FOXO3 and TRIM22 through bi-luciferase reporter assay. In other words, the increased expression of TRIM22 in GR cells may be due to the decreased expression of FOXO3 to some extent. Meanwhile, FOXO3 was considered to be one of the classic modulators of autophagy (23,24) . We have reasons to believe that, in vivo, autophagy activation by TRIM22 in lung cancer trigger by GEM may also be due to dysregulated FOXO3 expression. Unfortunately, due to the relationship between time and funds, this study cannot explore this issue.
Conclusions
The expression of TRIM22 was significantly upregulated in GR lung adenocarcinoma cell line A549. To some extent, it was due to the decreased expression of FOXO3, which is a transcription inhibitor of TRIM22. Due to the enhancement of pro-survival autophagy induced by TRIM22, the A549 cells became less sensitive to GEM to a certain degree.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The collection process of clinical serum samples involved in this research was approved and executed strictly in accordance with rules and regulations of the ethics committee of the Second Xiangya Hospital of Central South University (Ethical approval ID: EA20190001). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forster M, Hackshaw A, De Pas T, et al. A phase I study of nintedanib combined with cisplatin/gemcitabine as first-line therapy for advanced squamous non-small cell lung cancer (LUME-Lung 3). Lung Cancer 2018;120:27-33. [Crossref] [PubMed]
- Chen Y, Huang Y, Chen DM, et al. RRM1 expression and the clinicopathological characteristics of patients with non-small cell lung cancer treated with gemcitabine. Onco Targets Ther 2018;11:5579-89. [Crossref] [PubMed]
- Bian WG, Zhou XN, Song S, et al. Reduced miR-363-3p expression in non-small cell lung cancer is associated with gemcitabine resistance via targeting of CUL4A. Eur Rev Med Pharmacol Sci 2019;23:649-59. [PubMed]
- Bao L, Jaramillo MC, Zhang Z, et al. Induction of autophagy contributes to cisplatin resistance in human ovarian cancer cells. Mol Med Rep 2015;11:91-8. [Crossref] [PubMed]
- Nguyen HG, Yang JC, Kung HJ, et al. Targeting autophagy overcomes Enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene 2014;33:4521-30. [Crossref] [PubMed]
- Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem 2014;289:17163-73. [Crossref] [PubMed]
- Mani J, Vallo S, Rakel S, et al. Chemoresistance is associated with increased cytoprotective autophagy and diminished apoptosis in bladder cancer cells treated with the BH3 mimetic (-)-Gossypol (AT-101). BMC Cancer 2015;15:224. [Crossref] [PubMed]
- Button RW, Roberts SL, Willis TL, et al. Accumulation of autophagosomes confers cytotoxicity. J Biol Chem 2017;292:13599-614. [Crossref] [PubMed]
- Antunes F, Erustes AG, Costa AJ, et al. Autophagy and intermittent fasting: the connection for cancer therapy? Clinics (Sao Paulo) 2018;73:e814s. [Crossref] [PubMed]
- Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer 2017;36:52. [Crossref] [PubMed]
- Kim M, Jung JY, Choi S, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy 2017;13:149-68. [Crossref] [PubMed]
- Liu L, Zhou XM, Yang FF, et al. TRIM22 confers poor prognosis and promotes epithelial-mesenchymal transition through regulation of AKT/GSK3beta/beta-catenin signaling in non-small cell lung cancer. Oncotarget 2017;8:62069-80. [PubMed]
- Tooker P, Yen WC, Ng SC, et al. Bexarotene (LGD1069, Targretin), a selective retinoid X receptor agonist, prevents and reverses gemcitabine resistance in NSCLC cells by modulating gene amplification. Cancer Res 2007;67:4425-33. [Crossref] [PubMed]
- Lou J, Wang Y, Zheng X, et al. TRIM22 regulates macrophage autophagy and enhances Mycobacterium tuberculosis clearance by targeting the nuclear factor-multiplicity kappaB/beclin 1 pathway. J Cell Biochem 2018;119:8971-80. [Crossref] [PubMed]
- Zhu L, Huang F, Wan T, et al. Overexpression of long noncoding RNA LINC00882 is associated with poor prognosis in hepatocellular carcinoma. Onco Targets Ther 2018;11:5209-17. [Crossref] [PubMed]
- Liu Y, Wang X, Zhang Z, et al. The overexpression of Rab9 promotes tumor progression regulated by XBP1 in breast cancer. Onco Targets Ther 2019;12:1815-24. [Crossref] [PubMed]
- Jiang Y, Han Z, Wang Y, et al. Depletion of SIRT7 sensitizes human non-small cell lung cancer cells to gemcitabine therapy by inhibiting autophagy. Biochem Biophys Res Commun 2018;506:266-71. [Crossref] [PubMed]
- Huang XL, Zhang H, Yang XY, et al. Activation of a c-Jun N-terminal kinase-mediated autophagy pathway attenuates the anticancer activity of gemcitabine in human bladder cancer cells. Anticancer Drugs 2017;28:596-602. [Crossref] [PubMed]
- Wu HM, Shao LJ, Jiang ZF, et al. Gemcitabine-Induced Autophagy Protects Human Lung Cancer Cells from Apoptotic Death. Lung 2016;194:959-66. [Crossref] [PubMed]
- Wu HM, Jiang ZF, Ding PS, et al. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Sci Rep 2015;5:12291. [Crossref] [PubMed]
- Zhang X, Sai B, Wang F, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer 2019;18:40. [Crossref] [PubMed]
- Xiao K, Jiang J, Guan C, et al. Curcumin induces autophagy via activating the AMPK signaling pathway in lung adenocarcinoma cells. J Pharmacol Sci 2013;123:102-9. [Crossref] [PubMed]
- Fitzwalter BE, Thorburn A. FOXO3 links autophagy to apoptosis. Autophagy 2018;14:1467-8. [Crossref] [PubMed]
- Fitzwalter BE, Towers CG, Sullivan KD, et al. Autophagy Inhibition Mediates Apoptosis Sensitization in Cancer Therapy by Relieving FOXO3a Turnover. Dev Cell 2018;44:555-65.e3. [Crossref] [PubMed]


