
microRNA-324 plays an oncogenic role in bladder cancer cell growth and motility
Introduction
Bladder cancer (BC) is the ninth most common cancer worldwide, with an annual incidence of approximately 430,000 cases, and it is the 13th leading cause of cancer mortality (1). BC exhibits a male predominance and is the seventh most common cancer worldwide in men. The development of this cancer is significantly associated with exposure to environmental carcinogens. Because of the numerous industrial communities and the unique dietary culture associated with Chinese medicine, the molecular etiology of BC in Taiwanese patients is considerably different from that of BC in patients in other countries (2). The current therapy for BC demonstrates promising efficacy, with a 5-year survival rate of >60% (3). However, in some patients, the tumors are resistant to treatment and continue to grow and progress, leading to aggressive/invasive tumors, the treatment of which is challenging using the current treatment strategies (3). By combining the results of analysis of the genesis and evolution of cancer with functional validations of key drivers, in vitro genetic assays can be developed for predicting cancer recurrence and progression in patients (4).
microRNAs are small, nonprotein coding RNA transcripts and play a crucial role in modulating biological functions of human cancers, including BC (5,6). Our previous studies have indicated that the arm selection preference of microRNA varies in different types of human cancer cells during cancer progression (7-10). The expression of miR-324-5p or miR-324-3p suppressed the growth and invasion of breast cancer cells. Furthermore, ectopic expression of miR-324-5p reduced the growth and invasion abilities of colorectal cancer cells, whereas miR-324-3p suppressed colorectal cancer cell invasion but did not influence cell growth (8). Taken together, both miR-324-5p and miR-324-3p play a tumor suppressive role in breast and colon cancers. Notably, we reported the oncogenic role of miR-324-5p/-3p in lung cancer, revealing that miR-324-5p expression could promote cell growth and motility in lung cancer cells, whereas miR-324-3p overexpression could accelerate cell proliferation but not alter the invasion ability of lung cancer cells (10). However, the detailed roles of miR-324-5p and miR-324-3p in BC remain unclear. In this study, we investigated whether the overexpression of miR-324-5p and miR-324-3p and ectopic expression of miR-324-5p/-3p accelerate BC cell proliferation and invasion ability.
Methods
Analysis of expression data of miR-324 from The Cancer Genome Atlas (TCGA)
Small RNA sequence data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). A total 435 miRNA level-3 data, including those of 19 normal and 416 tumor tissues, were obtained from the TCGA data portal. Furthermore, the corresponding clinical information was obtained. The correlation between clinical features and miR-324-5p or miR-324-3p expression was further analyzed. In addition, the RNA profiles of 408 and 19 BC and adjacent normal tissues, respectively, were obtained from TCGA database.
Cell line
Human BC cell lines, BFTC950, were obtained from the American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% inactivated fetal bovine serum (Invitrogen, Carlsbad, CA, USA).
RNA extraction
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the instruction manual. The detailed procedure is described in our previous study (8). Thereafter, concentration, purity, and amount of total RNA were evaluated using a Nanodrop 1000 spectrophotometer (Nanodrop Technologies Inc., Wilmington, DE, USA).
Clinical samples
A total of 22 BC and 13 corresponding adjacent normal bladder tissue samples were collected from BC patients who underwent surgery at the Department of Surgery, Kaohsiung Veterans General Hospital, Kaohsiung, Taiwan. Our study protocol was independently reviewed and approved by the Institutional Review Board of Kaohsiung Veterans General Hospital (IRB approval number: VGHKS17-CT4-18).
Stem-loop reverse transcription (RT) and real-time polymerase chain reaction (PCR)
A total of 1 µg of total RNA was reverse-transcribed using stem-loop RT primers. Thereafter, real-time PCR was performed using a miR-324-5p- and miR-324-3p-specific forward primer and a universal reverse primer. The detailed procedure is described in our previous study (8). In the current study, the expression levels of miR-324-5p or miR-324-3p were normalized to those of U6. The following primer sequences were used to examine miRNAs:
miR-324-5p-RT: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACACCAAT-3'
miR-324-5p-GSF: 5'-CGGCGGCGCATCCCCTAGGGCAT-3'
miR-324-3p-RT: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCAGCAGC-3'
miR-324-3p-GSF: 5'- CGGCGGACTGCCCCAGGTGC-3'
Universal reverse: 5'- CTGGTGTCGTGGAGTCGGCAATTC-3'
U6-F: 5'-CTCGCTTCGGCAGCACA-3'
U6-R: 5'-AACGCTTCACGAATTTGCGT-3'
Ectopic expression of miR-324-5p and miR-324-3p with mimic transfection
BC cells, BFTC950, were transfected with 10 nM miRNA-324-5p mimics, miR-324-3p mimics, or a scrambled sequence as negative control (GenDiscovery Biotechnology Inc., Taiwan) by using the Lipofectamine RNAiMAX reagent (Invitrogen). After transfection for 48 h, the expression levels of miR-324-5p or miR-324-3p were assessed using stem-loop RT-qPCR.
Cell growth assay
In an assay to evaluate the colony formation ability, 2000 cells were seeded in a 6-well plate and then transfected with 10 nM miRNA-324-5p mimics, miRNA-324-3p mimics, or a scrambled control. Transfected cells were cultured for 2 weeks until the formation of colonies with substantial sizes. The colonies were stained using crystal violet solution and then dissolved in 1 mL of 10% acetic acid. The absorbance of individual wells was determined at 595 nm by using Multiskan FC (Thermo Scientific, USA). In the cell proliferation assay, 1,000 cells were seeded in a 96-well pate and then transfected with 10 nM miRNA-324-5p mimics, miRNA-324-3p mimics, or a scrambled control. The cell proliferation was evaluated at various time points (0, 1, 2, and 3 days) by using a CellTiter-Glo One Solution Assay (Promega Corporation, Madison, WI, USA). The detailed procedure is described in a previous study (11). All experiments were repeated three times.
Cell migration and invasion assays
The migration and invasion abilities of cells were assessed in vitro by using the transwell assay. The cells were seeded on the upper chamber of the transwells (Falcon, Corning Incorporated, USA) without Matrigel (BD Biosciences, MA, USA) for the migration assay or with a Matrigel coating for the invasion assay. The detail procedures are described in a previous study (11). All experiments were repeated three times.
Statistical analysis
The expression levels of miR-324-5p or miR-324-3p in BC tissues compared with those in their corresponding adjacent normal tissues from the TCGA database were analyzed using Student’s t tests. All experiments for cell growth, migration, and invasion were conducted in triplicate. Histograms present the mean values, and the error bars indicate the SD. These data were analyzed using Student’s t tests. The Mann–Whitney U test and Kruskal–Wallis one-way analysis of variance were used for comparing the expression levels of miR-324-5p or miR-324-3p between different clinicopathological features of patients with BC. In addition, the log-rank test and Cox proportional hazard model were used to evaluate the effects of different expression levels of miR-324-5p or miR-324-3p on BC patients’ OS and development of multiple primaries. The difference was considered significant at P<0.05.
Results
High miR-324-5p expression was strongly correlated with poor survival of patients with BC
Our previous studies have indicated that miR-324-5p and miR-324-3p play a tumor suppressive role in breast cancer and colon cancer (8). By contrast, miR-324-5p and miR-324-3p are significantly overexpressed in lung cancer cells, which accelerated the lung cancer cell growth and invasion ability on lung cancer cell growth and invasion ability (10). To elucidate the role of miR-324 in BC, we assessed its clinical impacts by analyzing data from TCGA database. Furthermore, we explored the biological function of miR-324 in BC cells by using a gain-of-function approach. We downloaded the expression data of 416 patients with BC and 19 corresponding adjacent normal tissues from the TCGA database. As shown in Figure 1, the expression levels of miR-324-5p and miR-324-3p significantly increased in BC tissues compared with those in the corresponding adjacent normal tissues (P=0.002 and P<0.001, respectively). In addition, we examined the expression levels of miR-324-5p and miR-324-3p in BC cells and adjacent normal tissues by using real-time PCR. Data revealed that miR-324-5p was significantly overexpressed in BC tissues compared with the corresponding adjacent normal tissues (P=0.037), whereas the expression of miR-324-3p exhibited no significant change in BC tissue (Figure 1C,D). We further downloaded the clinical information of 414 patients with BC from the TCGA database. We analyzed the clinical impacts of miR-324-5p and miR-324-3p expression on BC and found that high expression levels of miR-324-5p were associated with the pathological stage of cancer, whereas no significant correlation was observed between miR-324-3p expression and clinicopathological features (Table 1). We further categorized BC patients into two groups, high miR-324-5p or miR-324-3p expression group and low miR-324-5p or miR-324-3p expression group, by using the receiver operator characteristic curve. Kaplan–Meier analysis revealed that high miR-324-5p or miR-324-3p expression levels were borderline significantly correlated with the overall survival (OS) curve (miR-324-5p on OS: P=0.053 and miR-324-3p on OS: P=0.058; Figure 2, Table 2). Multivariate Cox regression revealed a significant association between high miR-324-5p expression levels and poor OS [adjusted HR (aHR) 1.65, 95% CI, 1.10–2.47, P=0.016; Table 2]. In addition, a significant association was observed between high miR-324-3p expression levels and poor OS (aHR 1.71, 95% CI, 1.03–2.84, P=0.039; Table 2). Our data indicated that high expression levels of miR-324-5p are an independent biomarker of the poor survival of BC patients.
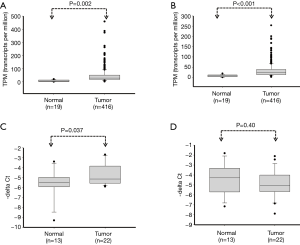
Table 1
| Variables | No. (%) | miR-324-5p (n=414) | miR-324-3p (n=414) | |||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | P value | Mean ± SD | Median | P value | |||
| Pathology stage (n=414) | ||||||||
| I | 4 (1.0) | 39.33±32.07 | 25.99 | <0.001b | 38.94±21.81 | 32.78 | 0.726a | |
| II | 131 (31.6) | 55.87±60.90ef | 37.09 | 40.28±37.80 | 30.86 | |||
| III | 141 (34.1) | 36.38±35.10e | 24.80 | 31.81±33.32 | 19.53 | |||
| IV | 138 (33.3) | 40.80±53.73f | 25.93 | 35.46±93.50 | 18.06 | |||
| pT stage (n=381) | ||||||||
| T1 | 5 (1.3) | 49.70±36.17 | 32.53 | 0.086a | 38.40±18.93 | 35.30 | 0.208a | |
| T2 | 120 (31.5) | 49.70±62.32 | 33.62 | 43.20±101.20 | 27.77 | |||
| T3 | 196 (51.4) | 37.48±42.47 | 24.78 | 29.71±30.32 | 18.13 | |||
| T4 | 60 (15.7) | 33.54±28.04 | 25.23 | 26.35±22.06 | 17.54 | |||
| pN stage (n=370) | ||||||||
| N0 | 238 (64.3) | 42.77±47.06 | 29.52 | 0.267a | 34.22±33.91 | 24.87 | 0.149a | |
| N1 | 47 (12.7) | 45.86±74.80 | 26.32 | 50.38±156.16 | 19.61 | |||
| N2 | 77 (20.8) | 33.18±34.26 | 24.38 | 24.42±23.68 | 15.13 | |||
| N3 | 8 (2.2) | 61.43±53.52 | 42.85 | 48.20±36.30 | 39.47 | |||
| pM stage (n=212) | ||||||||
| M0 | 201 (94.8) | 49.24±61.25 | 28.79 | 0.583c | 43.08±82.17 | 28.57 | 0.844c | |
| M1 | 11 (5.2) | 59.62±56.01 | 29.20 | 38.16±30.54 | 36.78 | |||
a, P values were estimated by one-way ANOVA test; b, P values were estimated by Kruskal-Wallis 1-way ANOVA test; c, P values were estimated by Mann-Whitney U test; d, P values were estimated by student’s t test; e, pathology stage II compared with III with statistically significant P<0.001; f, pathology stage II compared with IV with statistically significant P<0.001.
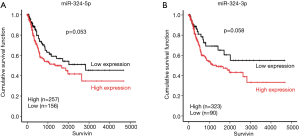
Table 2
| Characteristic | No. (%) (n=413) | Overall survival | |||
|---|---|---|---|---|---|
| CHR (95% CI) | P value | AHR (95% CI) | P value | ||
| miR-324-5p | |||||
| Low | 156 (37.8) | 1.00 | 1.00 | ||
| High | 257 (62.2) | 1.49 (0.99–2.22) | 0.055 | 1.65 (1.10–2.47) | 0.016 |
| miR-324-3p | |||||
| Low | 90 (21.8) | 1.00 | 1.00 | ||
| High | 323(78.2) | 1.63 (0.98–2.70) | 0.060 | 1.71 (1.03–2.84) | 0.039 |
AHR were adjusted for AJCC pathological stage (II, III and IV VS. I). CHR, crude hazard ratio; AHR, adjusted hazard ratio.
miR-324-5p and miR-324-3p play oncogenic roles in BC
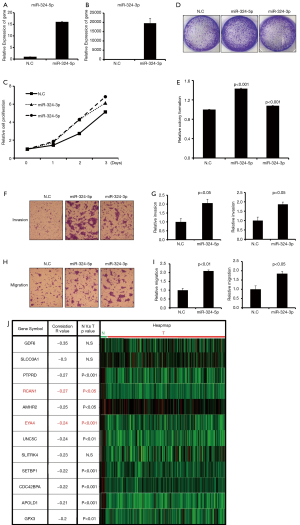
To investigate the biological function of the individual arms of miR-324, miR-324-5p and miR-324-3p mimics were respectively transfected into BFTC905. As shown in Figure 3A,B, miR-324-5p and miR-324-3p were obviously overexpressed in BFTC905 cells after individual mimic transfection, respectively. After transient transfection with miR-324-5p, miR-324-3p mimics, or scramble control, we further examined the biological functions, including cell proliferation, colony formation ability, and migration and invasion abilities. Our results indicated that ectopic miR-324-5p or miR-324-3p expression significantly accelerated BC cell growth, colony formation ability, and migration and invasion abilities in BFTC905 cells, especially miR-324-5p (Figure 3C,D,E,F,G,H,I). Therefore, we further illustrated target genes of miR-324-5p by bioinformatics approach. Using the TargetScan prediction tool, we identified that the 3'UTR region of 151 putative target genes may be targeted by miR-324-5p. In addition, the small RNA profiles and RNA profile, which contains 408 BCs and 19 adjacent normal tissues from the TCGA database, was downloaded. We analyzed the correlation and expression levels between miR-324-5p and 151 putative target genes in BC by analyzing TCGA database (Figure 3J). Because the expression levels of miR-324-5p were increased in BC, those putative targets candidates should be negative correlated with miR-324-5p expression and significantly decreased expressions in BC. According to the aforementioned criteria, we identified 9 putative target genes might be regulated by miR-324-5p in BC, including PTPRD, RCAN1, AMHR2, EYA4, UNC5C, SETBP1, CDC42BPA, APOLD1 and GPX3 (Figure 3J). Among them, previous studies reported that RCAN1 and EYA4 as tumor suppressors to inhibit BC growth and metastasis (12,13). In summary, miR-324-5p function as an oncogene to accelerate BC proliferation and invasion ability might be through directly regulating RCAN1 or EYA4 expression.
Discussion
Our previous studies have reported that the -5p and -3p arm of miRNA might have similar or antagonistic biological functions, depending on their target genes (7-10). Moreover, our previous study revealed that two mature miR-324 arms, miR-324-5p and miR-324-3p, were derived from pre-miR-324 during the maturation process, but the arm selection preference varies in different human cancer cells (8). Furthermore, the expression levels of miR-324-5p or miR-324-3p were simultaneously increased in some human cancer cell types, including BC. By contrast, miR-324-5p and miR-324-3p expression levels were decreased in colon cancer. Until now, most studies have reported that miR-324 plays a tumor suppressive role during cancer progression. Recently, we reported that miR-324-5p expression suppressed the growth and invasion abilities of colon cancer cells, whereas miR-324-3p expression inhibited colon cancer cell invasion but did not influence cell growth (8). In nasopharyngeal cancer, miR-324-3p expression suppressed cell growth and promoted apoptosis by silencing SMAD7 (14). In glioma cancer, miR-324-5p expression inhibited cancer cell growth by targeting GLI1 expression (15). In hepatocellular carcinoma (HCC), miR-324-5p expression was involved in the reduction of the migration and invasion capacity through the modulation of MMP2, MMP9, ETS1, and SP1 gene expression in HCC (16). Tang et al. reported that miR-324-5p is essential for multiple myeloma (MM) pathogenesis, and downregulation of miR-324-5p is a novel mechanism of hedgehog signaling activation in MM (17). Taken together, most studies have indicated that miR-324-5p and miR-324-3p play a tumor suppressive role in human cancer cells.
Although the expression levels of miR-324-5p and miR-324-3p are simultaneously increased in several human cancer types, including BC, breast cancer, head and neck cancer, lung cancer, and gastric cancer (8), few studies have indicated the oncogenic role of miR-324-5p or miR-324-3p in the modulation of cancer cell growth and motility. Tuo et al. reported that miR-324-3p promoted tumor growth by targeting DACT1 and activating the Wnt/βcatenin pathway in HCC (18). In addition, miR-324-3p promoted gastric cancer by activating the Smad4-mediated Wnt/beta-catenin signaling pathway (19). Lin et al. reported that both miR-324-5p and miR-324-3p expression accelerated lung cancer cell growth (10). Therefore, the role of miR-324 remains controversial in human cancer, and few studies have examined the biological role of miR-324-5p or miR-324-3p in BC. In the current study, we examined the biological function of miR-324-5p and miR-324-3p in BC and found that miR-324-5p and miR-324-3p expression significantly accelerated BC growth and invasion ability.
Accumulating evidence has revealed that miR-324 could be used as a diagnostic or prognostic biomarker of human cancer (20-23). Numerous studies have revealed that dysregulated miR-324 levels have been observed in the plasma of cancer patients. A study suggested that plasma miR-324-3p and miR-1285 levels may serve as lung cancer early detection markers, whereas miR-324-3p may serve as a prognostic marker of lung cancer (24). Jin et al. study revealed that the increased expression of serum miR-324 in patients with prostate cancer can serve as a biomarker for diagnosis (20). Wen et al. reported that the plasma miR-324-3p level was significantly increased in HCC, together with miR-20a-5p and miR-320a levels, which could serve as a diagnostic biomarker for the early detection of HCC (25). The expression of miR-185-3p and miR-324-3p was significantly reduced after RT in radioresistant but not in radiosensitive cases. Low miR-185-3p and miR-324-3p levels might be important markers for the prediction of low response to radiotherapy/chemoradiotherapy and poor OS and recurrence-free survival (14). In papillary thyroid cancer, miR-324-5p serves as a candidate predictor in addition to ultrasonography in predicting lymph node metastasis, especially central lymph node metastasis of unifocal papillary thyroid microcarcinoma without extracapsular spread (26).
Conclusions
Herein, our data indicated that high expression levels of miR-324-5p and miR-324-3p may serve as independent prognostics markers to predict the poor OS of patients with BC. Our findings indicated that miR-324 may be a promising prognostic biomarker and may provide a new insight into the therapy of BC.
Acknowledgments
The authors appreciated the assistance of Biobank, Department of Medical Education and Research, Kaohsiung Veterans General Hospital, for the processing of clinical specimen.
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Kaohsiung Veterans General Hospital (IRB approval number: VGHKS17-CT4-18). The methods were carried out in accordance with the approved guidelines and all patients provided informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Yeh HL, Hsu SW, Chang YC, et al. Spatial Analysis of Ambient PM2.5 Exposure and Bladder Cancer Mortality in Taiwan. Int J Environ Res Public Health 2017; [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta 1994;1198:11-26. [PubMed]
- Liu X, Wu Y, Zhou Z, et al. Celecoxib inhibits the epithelial-to-mesenchymal transition in bladder cancer via the miRNA-145/TGFBR2/Smad3 axis. Int J Mol Med 2019;44:683-93. [PubMed]
- Liang T, Wang Y, Wang Y, et al. Long noncoding RNA PEG10 facilitates bladder cancer cells proliferation, migration, and invasion via repressing microRNA-29b. J Cell Physiol 2019;234:19740-9. [Crossref] [PubMed]
- Tsai KW, Leung CM, Lo YH, et al. Arm Selection Preference of MicroRNA-193a Varies in Breast Cancer. Sci Rep 2016;6:28176. [Crossref] [PubMed]
- Kuo WT, Yu SY, Li SC, et al. MicroRNA-324 in Human Cancer: miR-324-5p and miR-324-3p Have Distinct Biological Functions in Human Cancer. Anticancer Res 2016;36:5189-96. [Crossref] [PubMed]
- Chou NH, Lo YH, Wang KC, et al. MiR-193a-5p and -3p Play a Distinct Role in Gastric Cancer: miR-193a-3p Suppresses Gastric Cancer Cell Growth by Targeting ETS1 and CCND1. Anticancer Res 2018;38:3309-18. [Crossref] [PubMed]
- Lin MH, Chen YZ, Lee MY, et al. Comprehensive identification of microRNA arm selection preference in lung cancer: miR-324-5p and -3p serve oncogenic functions in lung cancer. Oncol Lett 2018;15:9818-26. [PubMed]
- Tsai KW, Kuo WT, Jeng SY. Tight Junction Protein 1 Dysfunction Contributes to Cell Motility in Bladder Cancer. Anticancer Res 2018;38:4607-15. [Crossref] [PubMed]
- Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer 2019;18:95. [Crossref] [PubMed]
- Eissa S, Safwat M, Matboli M, et al. Measurement of Urinary Level of a Specific Competing endogenous RNA network (FOS and RCAN mRNA/ miR-324-5p, miR-4738-3p, /lncRNA miR-497-HG) Enables Diagnosis of Bladder Cancer. Urol Oncol 2019;37:292 e19- 27.
- Xu J, Ai Q, Cao H, et al. MiR-185-3p and miR-324-3p Predict Radiosensitivity of Nasopharyngeal Carcinoma and Modulate Cancer Cell Growth and Apoptosis by Targeting SMAD7. Med Sci Monit 2015;21:2828-36. [Crossref] [PubMed]
- Xu HS, Zong HL, Shang M, et al. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci 2014;18:828-32. [PubMed]
- Cao L, Xie B, Yang X, et al. MiR-324-5p Suppresses Hepatocellular Carcinoma Cell Invasion by Counteracting ECM Degradation through Post-Transcriptionally Downregulating ETS1 and SP1. PLoS One 2015;10:e0133074. [Crossref] [PubMed]
- Tang B, Xu A, Xu J, et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int J Cancer 2018;142:109-20. [Crossref] [PubMed]
- Tuo H, Wang Y, Wang L, et al. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/beta-catenin pathway in hepatocellular carcinoma. Oncotarget 2017;8:65687-98. [Crossref] [PubMed]
- Sun GL, Li Z, Wang WZ, et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J Gastroenterol 2018;53:725-39. [Crossref] [PubMed]
- Jin YY, Tong SQ, Tong M. Diagnostic Value of Circulating miR-324 for Prostate Cancer. Clin Lab 2019; [Crossref] [PubMed]
- Chang CC, Lin CC, Hsieh WL, et al. MicroRNA expression profiling in PBMCs: a potential diagnostic biomarker of chronic hepatitis C. Dis Markers 2014;2014:367157.
- Zhao JY, Wang F, Li Y, et al. Five miRNAs Considered as Molecular Targets for Predicting Esophageal Cancer. Med Sci Monit 2015;21:3222-30. [Crossref] [PubMed]
- Namkung J, Kwon W, Choi Y, et al. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol 2016;31:1160-7. [Crossref] [PubMed]
- Gao X, Wang Y, Zhao H, et al. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget 2016;7:59664-75. [PubMed]
- Wen Y, Han J, Chen J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer 2015;137:1679-90. [Crossref] [PubMed]
- Yang Y, Xia S, Ni X, et al. MiR-324-5p assists ultrasonography in predicting lymph node metastasis of unifocal papillary thyroid microcarcinoma without extracapsular spread. Oncotarget 2017;8:83802-16. [PubMed]


