
Chidamide inhibits t(8;21) AML cell proliferation and AMK1/ETO and C-KIT expression by inhibiting ERK1/2 signaling pathway
Introduction
Acute myeloid leukemia (AML) is a cloner disorder derived from the myeloid hematopoietic stem cell progenitors (1). Appropriately 60% of the AML patients are associated with cytogenetic abnormalities. t(8;21)(q22;q22) is the most common chromosomal translocation (2,3), which leads to AML1-ETO (acute myeloid leukemia1/myeloid transforming gene 8) fusion protein. AML1-ETO fusion is a transcription factor that inhibits the apoptosis of leukemia cells. t(8;21)AML is featured by high heterogeneity. Although some patients can achieve long-term survival through standard treatment regimen, 20–40% of patients can not achieve complete remission (CR) and 60% of patients relapse. To date, the reasons of this heterogeneity remains unknown. The treatment of t(8;21) AML has been the major challenge in the management of hematologic malignancies.
Epigenetic changes such as deacetylation and acetylation of histone play a crucial role in the occurrence and progression of tumor. AML1-ETO can interact with HDAC1, HDAC2 and HDAC3, inhibit normal transcription, promote the proliferation of tumor cells, reduce the normal differentiation of granulocytes and induce the apoptosis of leukemia cells (4). HDACi is a group of small-molecule compounds with extensive anti-tumor activity. It has been reported that HDACi can be used to treat t(8;21)AML. Several types of HDACi are currently under clinical trials for hematological tumor (5-7). Chidamide is the first HDACi that is independently developed in China, and it has been clinically used to treat the rare peripheral T cell lymphoma (PTCL) (8,9). However, the effect and mechanism of HDACi on t(8;21)AML are not yet clarified. In this study, we aimed to investigate the effect of chidamide on t(8;21) AML cells and explore its mechanism.
Methods
Reagents
Chidamide was obtained from Shenzhen Chipscreen and prepared into 2 mM stock solution using Dimethyl sulfoxide (DMSO). The solution was preserved at −20 °C and thawed prior to use. RIPM1640 medium was purchased from Hyclone (USA). Fetal bovine serum (FBS) was purchased from Gibco (USA). CCK-8 was purchased from Dojindo (Japan). Annexin V-FITC apoptosis detection kit was manufactured by Dojindo. Anti-histone H3 antibody, anti-acetyl-histone H3 antibody, anti-p44/42MAPK (ERK1/2) antibody, and anti-Pp44/42MAPK (ERK1/2) antibody were purchased from Cell Signaling (USA). Anti-AML1/RHD antibody was purchased from Calbiochem (USA). Anti-C-KIT antibody, β-acting, and HRP-labeled secondary antibodies were purchased from Santa Cruz (USA). Lymphocyte separation medium (Ficoll) was purchased from GE Healthcare (USA). Trizol was purchased from Ambion (USA). Reverse transcription kit was purchased from Promega (USA). KAPA SYBR FAST q-PCR Master Kit was purchased from KAPA (USA).
Cell lines and cell culture
Human AML cell lines Kasumi-1 and SKNO-1, human promyelocytic leukemia cell line HL-60, and human acute monocytic leukemia cell line U937 were purchased from American Type Culture Collection (ATCC). U937 cells were transfected with AML1/ETO fusion gene to obtain U937-A/E. AML1/ETO-siRNA was transfected to SKNO-1 cells to obtain SKNO-1-si-A/E cells. AML cells were cultured in RIPM1640 medium containing 10% FBS and 1% penicillin-streptomycin at 37°C in a humidified incubator with 5% CO2. Cells were subcultured every two to three days. Exponential-phase cells were harvested for further experiments.
Cell proliferation assay
The inhibitory effect of chidamide on the proliferation of AML cells was detected by CCK-8 assay. Exponential-phase cells were inoculated into a 96-well plate at a density of 1.0×103–1.0×104 cells per well. In different treatment groups, 0.125, 0.25 and 0.5 µM chidamide was added, respectively. An equal amount of DMSO was added in control group. A blank group containing culture medium without cells was set up. Each group had three replicates. After incubation for 24–144 h, 10 µL of CCK-8 was added into each well in the dark and the cells were incubated for an additional 3 h. The absorbance (A) at 450 nM was measured using a microplate reader. The results were zeroed using the blank group. The inhibition rate was calculated as follows: inhibition rate (%) = [(A of control group-A of blank group) − (A of experimental group-A of blank group)]/[A of control group-A of blank group] ×100%. The experiment was repeated three times.
Flow cytometry
Exponential-phase AML cells were inoculated into a 24-well plate at a density of 2×104 cells per well, for 1ml in each well. In different treatment groups, 0.25 and 0.5 µM chidamide was added into each well, respectively. In control group, an equal amount of DMSO was added. Each group had three replicates. After 48 or 96 h, cells were harvested and washed twice with pre-cooled PBS. Cells were resuspended in 1X binding buffer, stained with 5 µL of FITC Annexin V and 10 µL of propidium iodide (PI) in the dark for 10 min. The fluorescence of cells was detected using a BD LSRFortessa™ flow cytometer (BD Biosciences) at 488 nm within 1 h and analyzed using ModFit LT software.
Cell cycle experiment
Exponential-phase AML cells were inoculated into a 6-well plate at a density of 2×104 cells per well. In different treatment groups, 0.25 and 0.5 µM chidamide was added into each well, respectively. In control group, an equal amount of DMSO was added. After 48 h, cells were collected at 180× g for 5 min, washed twice with PBS, fixed in pre-cooled 70% ethanol and digested by 50 µg/mL RNase. Cells were stained with 50 µg/mL PI in a 37 °C water bath for 30 min. The percentage of cell population at S, G0/G1 or G2 phase was analyzed by flow cytometry.
Microarray profiling
Kasumi-1 cells were treated with 0.25 µM chidamide for 48 h. Total RNA was extracted using Trizol reagent. Gene expression was detected using Phalanx Human One Array Ver. 6 Release 2. Data were processed using Rosetta error model. Repetitive probes were combined and the signals were normalized. Differential genes were defined by (I) and (II): (I) log2 |Fold change| ≥1 and P<0.05; (II) log2 |Fold change|= “NA”, the signal difference between two samples ≥1,000. Principal analysis, clustering analysis and functional enrichment analysis were applied to differential genes. The probes were annotated using NCBI, Gene Ontology, Ensembl and Uniprot.
Western blot analysis
Exponential-phase AML cells were inoculated into a 6-well plate at a density of 2×106 cells per well. In different treatment groups, 0.25 and 0.5 µM chidamide was added into each well, respectively. In control group, an equal amount of DMSO was added. Cells were cultured for 6, 12, 24, 48, 72 and 96 h, respectively. Proteins were extracted using radio-immunoprecipitation assay (RIPA) containing 1 mM PMSF (Phenylmethanesulfonyl fluoride). Protein concentrations were quantified by BCA assay. Proteins were separated by SDS-PAGE (SDS-polyacrylamide gel electrophoresis) and transferred to PVDF (hydrophobic polyvinylidene fluoride) membrane. The membrane was blocked with 5% defatted milk for 1 h, and incubated with primary antibodies overnight at 4 °C on a shaker. The membrane was washed three times with TBS-T (Tris-HCl buffered saline solution with Tween 20) and incubated with secondary antibody for 40 min. The membrane was then treated with hyper-sensitive ECL substrate and the color was developed in dark. β-actin was used as the internal reference.
Real time quantitative PCR (RT q-PCR) assay
Kasumi-1 and SKNO-1 cells were respectively inoculated into a 24-well plate at a density of 2×106 cells per well. In different treatment groups, 0.25 and 0.5 µM chidamide was added into each well, respectively. In control group, an equal amount of DMSO was added. MEK1/2-specific inhibitor U0126 (10 nM) was added to each well. After 48-h incubation, total RNA was extracted using Trizol reagent and reverse transcribed into cDNA at 42 °C. qPCR was performed using KAPA SYBR FAST q-PCR Master Kit in an ABI 7500 System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: 95 °C for 10 min followed by 35 cycles of 95 °C for 10 sec, 58 °C for 30 sec and 72 °C for 30 sec. Data was analyzed using the 2-ΔΔCt method. The relative expression of AML1/ETO and C-KIT was calculated using β-actin as the internal control.
Statistical analysis
Normality and homogeneity of variance were tested in each group. Normally distributed data were expressed as mean ± standard deviation (). t-test was used for difference between two groups. Univariate multi-level one-way ANOVA was used for multiple group comparison. Other data were expressed by median and analyzed by rank sun test. P<0.01 was considered extremely significant, and P<0.05 was considered significant. All statistical analyses were performed using SPSS 17.0 software.
Results
Chidamide inhibited the proliferation of AML cells
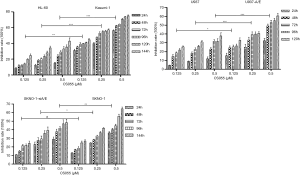
Chidamide inhibited the proliferation of AML cells in a time- and dose-dependent manner. The inhibitory effect was stronger in the AML1/ETO-positive Kasumi-1 and U937-A/E cells compared with AML1/ETO-negative HL-60 and U937 cells (P<0.001, Figure 1). Moreover, the inhibitory effect of 0.5 µM chidamide was significant higher compared with 0.25 µM chidamide (P<0.001). 0.125 µM chidamide had similar inhibitory effect on SKNO-1 and SKNO-1-siA/E cells, whereas 0.25 and 0.5 µM chidamide had stronger inhibitory effects on SKNO-1 cells compared with SKNO-1-siA/E cells. Thus 0.25 and 0.5 µM chidamide were used in further experiments.
Chidamide induced the apoptosis of AML1/ETO-positive AML cells
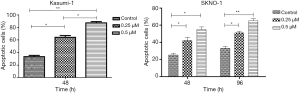
To find out the mechanism of proliferation inhibition by chidamide, the effects of chidamide on the apoptosis of AML1/ETO positive cells was examined. 0.25 and 0.5 µM chidamide significantly induced the apoptosis of Kasumi-1 and SKNO-1 cells (Figure 2), but had little effect on U937 and SKNO-1-siAE cells.
Chidamide induced cell cycle arrest in t(8;21)AML cells
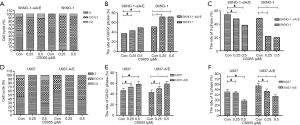
Cell cycle arrest might be another mechanism of cell proliferation inhibition by chidamide, and therefore was also detected. In AML1/ETO-positive cells, chidamide treatment induced a significant increase in the proportion of G0/G1 and G2 phase cells, and a significant decrease in the proportion of S phase cells (Figure 3). However, chidamide had not affected the percentage of G0/G1, G2 or S phase cells in AML1/ETO-negative cells, indicating that it could not induce cell cycle arrest in these cells.
Microarray gene expression profiling in AML1/ETO-positive AML cells after chidamide treatment
To study further the acting mechanism of chidamide, microarray gene expression profiling was performed to detect the functions of different genes. We first summarized the molecular functions by GO analysis. “apoptosis” related genes were changed impressively (P<0.05). It is surprise to note that “MAPKKK cascade” related genes were also involved (Figure 4).
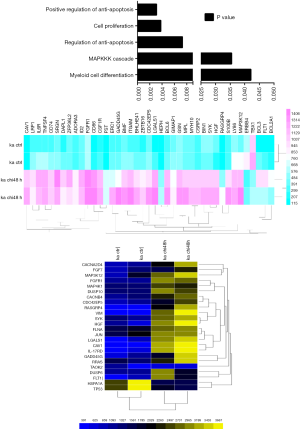
MAPK-related genes are involved in the activation of MAPK (mitogen-activated protein kinase) signaling pathway, indicating that chidamide might work through MAPK signaling pathway. Thus the MAPK-related genes with significant expression changes (log2 value ±1 in the chidamide treatments compared with the control) were further analyzed. These differentially expressed genes were associated with cell apoptosis, differentiation and cell cycle regulation. Most of them are regulated by three members of MAPK family: ERK1/2, p38-MAPK and JNK1/3 (c-Jun amino-terminal kinase 1/3). p38-MAPK and JNK1-3 are mainly involved in inflammatory responses, and ERK1/2 play an important role in the regulation of cell proliferation and differentiation (Figure 5).
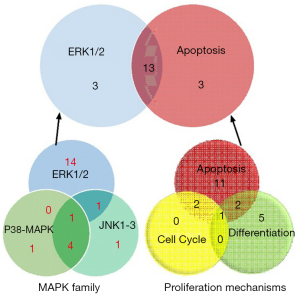
By analyzing the three important family members in MAPK pathway, the numbers of differentially expressed genes were studied. Sixteen “ERK1/2” related genes were changed among the total 22 “MAPK” family related genes, while 16 “apoptosis” and 3 “cell cycle” related genes were grouped by the proliferation mechanisms. Intriguingly, 13 genes were inclusive both in the “ERK1/2” pathway and “apoptosis” functions (Figure 5). These results indicated that chidamide might inhibit the ERK pathway by promoting the apoptosis of AML1/ETO-positive cells.
Chidamide inhibited histone 3 acetylation and ERK1/2 phosphorylation in AML1/ETO-positive AML cells
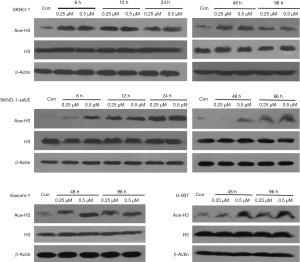
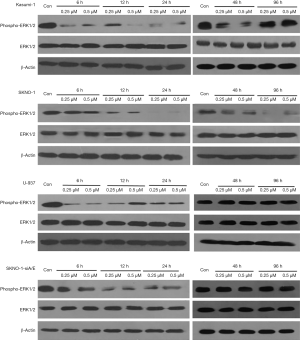
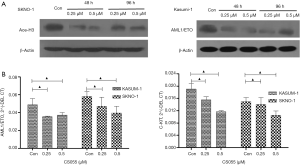
Chidamide is a selective HDAC inhibitor. Study (10) has shown that chidamide increases the levels of acetylated histone 3 (H3) and acetylated histone 4 (H4) in human B cell lymphoma cell lines. Recent reports are generally concerned about the effect of H3. Therefore, we focused on the effect of chidamide on H3 expression in AML cells. H3 was acetylated in AML1/ETO-positive AML cells (SKNO-1 and Kasumi-1) since 6 h after treatment with low-dose chidamide (0.25 µM) till 96 h, whereas in AML1/ETO-negative AML cells (SKNO-1-SiA/E and U937), 0.25 µM chidamide had not acetylated H3 at 6 or 48 h (Figure 6). When compared with AML1/ETO-negative AML cells (SKNO-1-SiA/E and U937), chidamide exhibited a higher and longer inhibitory effect on phospho-ERK1/2 in AML1/ETO-positive AML cells (96- and 48-h inhibition in SKNO-1 and Kasumi-1 cells, respectively). The expression of ERK1/2 remains constant (Figure 7). It has been reported that HDAC inhibitors can degrade AML1-ETO fusion protein via the ubiquitin-proteasome pathway (11). Nevertheless, the effect of chidamide on the expressions of AML1-ETO and C-KIT genes is not fully understood. Our results suggested that chidamide reduced the expression of AML1/ETO fusion protein and C-KIT protein in AML1/ETO-positive AML cells,but had not significantly affected AML1/ETO and C-KIT mRNA expression (Figure 8).
U0126 inhibited the proliferation of AML1/ETO-positive AML cells via regulating the ERK1/2 pathway
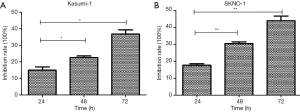
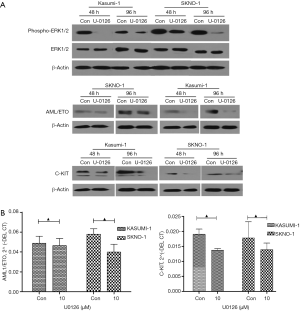
We further investigated whether chidamide decreased the proliferation of AML1/ETO-positive cells through inhibition of the ERK1/2 pathway. We used U0126, a special inhibitor of ERK1/2, to treat cells. Results showed that U0126 significantly inhibited the proliferation of AML1/ETO-positive cells (Kasumi-1 and SKNO-1) compared with the control group (Figure 9). U0126 treatment also downregulated the activation of phospho-ERK1/2, and reduced AML1/ETO and C-KIT protein expression in Kasumi-1 and SKNO-1 cells (Figure 10A). The AML1-ETO and C-KIT mRNA expression remained unchanged after U0126 treatment (Figure 10B).
Discussion
The prognosis of t(8;21) AML is usually good. Most patients can benefit from large-dose Ara-C (Cytarabine) chemotherapy and achieve remission after standard treatment regimen. However, an increasing number of studies have revealed high heterogeneity in t(8;21) AML, and approximately 40% of these patients are refractory or recurrent cases. Therefore, novel drugs or treatment are urgently needed for t(8;21) AML.
HDACi is an epigenetic regulator that can induce differentiation, cell cycle arrest, and apoptosis in tumor cells. HDACi has received an increasing attention in the anti-tumor field. HDACi can specifically inhibit AML1/ETO-positive AML cells by inducing apoptosis and differentiation. Moreover, HDACi can increase the acetylation level of histone H3 and H4. Chidamide is a selective HDACi independently developed in China, that can inhibit the proliferation, induce the apoptosis and cell cycle arrest of AML cells (12,13). We herein investigated the effect and mechanism of chidamide on t(8;21) AML cells. Our preliminary study has found that chidamide inhibits SKNO-1 xenograft tumor growth in nude mice (data not shown). In this study, three different concentrations of chidamide (0.125, 0.25 and 0.5 µM) were administered to AML cells to mimic the human blood concentration of chidamide. It was found that chidamide effectively inhibited the proliferation, decreased cell proportion in GO/G1 and S phase, and induced the apoptosis of Kasumi-1 and SKNO-1 cells. We further investigation the acting molecular mechanism of chidamide in AML1/ETO-positive AML cells. It is known that ETO and AML1-ETO react with the transcriptional corepressor and HDACs, indicating that the AML1-ETO recruitment of HDACs may play an important role in the pathogenesis of leukemia. C-KIT gene mutation is one of the most common mutations associated with t(8;21) AML, with an incidence of 17–46% (14). As an oncogene, C-KIT encodes for the receptor tyrosine kinase, and targets stem cell factor (SCF). The binding of C-KIT receptor to SCF regulates the proliferation of hematopoietic cells, as well as the malignant progression of tumor cells. Moreover, C-KIT works synergistically with AML1-ETO fusion protein to promote the conversion of hematopoietic stem cells towards leukemia. Therefore, C-KIT mutation predicts poor prognosis in t(8;21) AML (15). In this study, the effect of chidamide on C-KIT and AML-ETO protein expression was evaluated in AML1/ETO-positive AML cells carrying C-KIT mutation. It was found that chidamide reduced the expression of AML1/ETO and C-KIT in Kasumi-1 and SKNO-1 cells, but had no effects on mRNA expressions of either gene, indicating that chidamide did not regulate AML cells at the transcriptional level.
Microarray profiling of AML1/ETO-positive AML cells was performed to further explore the molecular mechanism of the inhibitory effect of chidamide on the proliferation of t(8;21) AML cells. Results suggested that chidamide might exert the regulatory role via MAPK signaling pathway. Further analysis revealed that most differentially expressed genes in MAPK pathway were related to ERK1/2 pathway, which is a key regulator for cell proliferation and differentiation. These results suggested that chidamide might promote the apoptosis of the AML1/ETO-positive AML cells by inhibiting the ERK1/2 pathway.
MAPK is a highly conservative serine/threonine protein kinase in the cytoplasm, which transmits extracellular signals to the interior of cells and thus regulates a diversity of biological functions (16). ERK (extracellular signal-regulated kinase), an important member of MAPK family, consists of two highly homologous subtypes, ERK1 and ERK2. ERK 1/2 is mainly involved in the Ras-Raf-MAPK signaling pathway, which is closely associated with leukemia. Abnormal activation of the ERK1/2 signaling pathway interrupts normal cell apoptosis and differentiation, and promotes malignant cell conversion (17). Recent study has shown that the ERK1/2 pathway is closely related to the sensitivity and tolerance of leukemia to chemotherapy (18). In this study, phospho-ERK1/2, the activated form of ERK1/2, was upregulated in t(8;21) AML cells after chidamide treatment. However, it remains unknown whether chidamide inhibits the proliferation of AML1/ETO-positive AML cells via the Ras/Raf/MEKERK pathway. We found that the acetylation level of histone H3 was increased over time. The expressions of phospho-ERK1/2 in AML1/ETO-positive AML cells was reduced after chidamide inhibition. We further examined whether other pathways were involved in the regulation of chidamide on AML1/ETO-positive AML cells. We found that U0126, a MEK1/2-specific inhibitor, significantly inhibited the proliferation of AML1/ETO-positive Kusumi-1 and SKNO-1 cells, and effectively downregulated the expression of phospho-ERK1/2 in both cells. All together, our results suggested that the inhibitory effect of chidamide on AML cells was achieved through modulating the ERK1/2 pathway.
In summary, we confirmed that chidamide inhibited the proliferation of t(8;21)AML cells by inducing apoptosis and cell cycle arrest. The working mechanism was related to the inhibition of phospho-ERK1/2 and oncogenes AML1/ETO and C-KIT. This finding sheds new light on the development of targeted therapy for t(8;21) AML patients, especially those with C-KIT mutation.
Acknowledgments
We are grateful to a Doctoral Innovation Fund that has been provided to Jing Liu by the Chinese PLA General Hospital.
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gyurkocza B, Lazarus HM, Giralt S. Allogeneic hematopoietic cell transplantation in patients with AML not achieving remission: potentially curative therapy. Bone Marrow Transplant 2017;52:1083-90. [Crossref] [PubMed]
- Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene 2001;20:5660-79. [Crossref] [PubMed]
- Mahmud H, Scherpen FJG, de Boer TM, et al. Peptide microarray profiling identifies phospholipase C gamma 1 (PLC-γ1) as a potential target for t(8;21) AML. Oncotarget 2017;8:67344-54. [Crossref] [PubMed]
- Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark Ed) 2012;17:1120-39. [Crossref] [PubMed]
- Issa JP, Garcia-Manero G, Huang X, et al. Results of phase 2 randomized study of low-dose decitabine with or without valproic acid in patients with myelodysplastic syndrome and acute myelogenous leukemia. Cancer 2015;121:556-61. [Crossref] [PubMed]
- Vey N, Prebet T, Thalamas C, et al. Phase 1 dose-escalation study of oral abexinostat for the treatment of patients with relapsed refractory higher-risk myelodysplastic syndromes, acute myeloid leukemia, or acute lymphoblastic leukemia. Leuk Lymphoma 2017;58:1880-6. [Crossref] [PubMed]
- Rücker FG, Lang KM, Fütterer M, et al. Molecular dissection of valproic acid effects in acute myeloid leukemia identifies predictive networks. Epigenetics 2016;11:517-25. [Crossref] [PubMed]
- Gu R, Liu T, Zhu X, et al. Development and validation of a sensitive HPLC-MS/MS method for determination of chidamide (epidaza), a new benzamide class of selective histone deacetylase inhibitor, in human plasma and its clinical application. J Chromatogr B Analyt Technol Biomed Life Sci 2015;1000:181-6. [Crossref] [PubMed]
- Xu Y, Zhang P, Liu Y. Chidamide tablets: HDAC inhibition to treat lymphoma. Drugs Today (Barc) 2017;53:167-76. [Crossref] [PubMed]
- Li YY, Wang YF, Wang J, et al. Effect of chidamide on human B lymphoma cell lines and its mechanisms. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2012;20:893-9. [PubMed]
- Chen LP, Zhang JW, Xu FM, et al. Dexamethasone and vorinostat cooperatively promote differentiation and apoptosis in Kasumi-1 leukemia cells through ubiquitination and degradation of AML1-ETO. Zhonghua Xue Ye Xue Za Zhi 2013;34:741-4. [PubMed]
- Gong K, Xie J, Yi H, et al. CS055 (Chidamide /HBI-8000), a novel histone deacetylase inhibitor, induces G1 arrest, ROS-dependent apoptosis and differentiation in human leukaemia cells. Biochem J 2012;443:735-46. [Crossref] [PubMed]
- Shi P, Zhang L, Chen K, et al. Low-dose decitabine enhances chidamide-induced apoptosis in adult acute lymphoblast leukemia, especially for p16-deleted patientsthrough DNA damage. Pharmacogenomics 2017;18:1259-70. [Crossref] [PubMed]
- Chen LT, Chen CT, Jiaang WT, et al. BPR1J373, an Oral Multiple Tyrosine Kinase Inhibitor, Targets c-KIT for the Treatment of c-KIT-Driven Myeloid Leukemia. Mol Cancer Ther 2016;15:2323-33. [Crossref] [PubMed]
- Ayatollahi H, Shajiei A, Sadeghian MH, et al. Prognostic Importance of C-KIT Mutations in Core Binding Factor Acute Myeloid Leukemia: A Systematic Review. Hematol Oncol Stem Cell Ther 2017;10:1-7. [Crossref] [PubMed]
- Cossa G, Gatti L, Cassinelli G, et al. Modulation of sensitivity to antitumor agents by targeting the MAPK survival pathway. Curr Pharm Des 2013;19:883-94. [Crossref] [PubMed]
- Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur J Med Chem 2016;109:314-41. [Crossref] [PubMed]
- Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017;8:59950-64. [PubMed]


