Potential of microRNA expression profile in predicting renal impairment risk in multiple myeloma patients
Introduction
Multiple myeloma (MM), as a hematological malignancy featured by clonal proliferation of malignant plasma cells which accounts for around 1% of neoplastic diseases and 13% of hematologic malignancies, commonly accompanied with severe symptoms such as renal impairment (RI), extensive skeletal destruction, infections, anemia, hypercalcemia and so on (1). Among these complications, RI occurs in 20–50% MM patients worldwide, while in China, RI is reported in 24.0%, 19.7% and 30.8% of total MM patients in China mainland, Hong Kong and Taiwan, respectively (2,3). Although bortezomib and dexamethasone triplet combinations are proposed to be the current standard of therapy for MM patients with RI (RI-MM patients), some patients refractory to these indications or with low-income are hard to benefit from the treatment, besides, the RI still greatly weakens the prognosis of MM patients (4,5). Thus, exploration of novel biomarkers for RI risk in MM patients is essential for the early prevention, treatment and prognosis improvement.
MicroRNA (miRNA), as a group of non-coding RNAs, has discloses its wide application as biomarkers for many diseases including renal diseases (6). For instance, a previous study reports that the miRNA expression pattern is correlated with the vascular remodeling in patients with type 2 diabetes mellitus (7); another study uncovers that abnormal miRNA expression profile is suggestive for renal fibrosis in patients with Fabry disease (8); besides, a meta-analysis reveals the potential of miRNA expression profile as biomarker for renal fibrosis (6). Apart from these evidences, miRNA expression profile is also discovered to be deeply involved in the development and progression of MM (9,10). Considering these above-mentioned data, we hypothesized that miRNA expression profile might have the potential to be biomarker for RI risk in MM patients, while no-related study has ever been reported.
Thus, this present study aimed to investigate the potential of miRNA expression profile in predicting RI risk in MM patients.
Methods
Patients
Between January 2016 and February 2019, 60 RI-MM patients and 60 MM patients without RI (non-RI-MM patients) treated in our hospital were recruited in this study. The inclusion criteria for RI-MM patients were as follows: (I) diagnosed as primary MM in accordance with the International Myeloma Working Group (IMWG) updated criteria for the diagnosis of MM (11); (II) presented with RI, which was defined as serum creatinine >1.73 µmol/L (or >2 mg/dL) or estimated creatinine clearance <40 mL/min and confirmed by the renal biopsy (11); (III) age ≥18 years. And the inclusion criteria for non-RI-MM patients were: (I) diagnosed as primary MM according to the IMWG criteria (11); (II) absence of RI confirmed by clinical and laboratory examinations; (III) age ≥18 years. Both the RI-MM patients and the non-RI-MM patients were excluded from the study if they had any of following conditions: (I) secondary or relapsed MM; (II) concomitant with other malignancies; (III) pregnant or lactating women. The present study was approved by the Institutional Review Board of our hospital with ethical approval number 2015-018, and all patients provided the written informed consents.
Data and sample collection
After confirmation of patients’ eligibility, clinical data and biochemical indexes were recorded prior to initial treatment, which mainly covered age, gender, body weight and the level of haemoglobin (Hb), calcium, serum creatinine (Scr), creatinine clearance rate (Ccr), albumin (ALB), β2 microglobulin (β2-MG) and lactate dehydrogenase (LDH). Before initial therapy, bone marrow samples were extracted from all patients, then the plasma cells were isolated from the samples using CD138+ Plasma Cell Isolation Kit (Miltenyi, Bergisch Gladbach, Germany) and stored at −80 °C for further determination.
Microarray
Twenty plasma cell samples from RI-MM patients and 20 plasma cell samples from non-RI-MM patients were subjected to microarray assay. In brief, total RNA was extracted from each sample by Trizol reagent (Invitrogen, Carlsbad, USA) followed by RNA integrity assessment by Agilent 2100 Bioanalyzer (Agilent, City of Santa Clara, USA) and quantification by NanoDrop ND-1000 spectrophotometer (Thermo, Wilmington, USA), Subsequently, miRNA expression profile of each sample was detected using Affymetrix Multispecies miRNA-4 Array (Agilent, City of Santa Clara, USA) by Genergy Bio (Shanghai, China) as the methods described in previous studies (12,13).
Bioinformatics
As for data processing, quantile normalization and low-intensity filtering were performed by R software package (R version 3.1.2), and the miRNAs detected in above 50 percent samples were proposed to further analysis. Bioinformatics analysis was performed using R software package (R version 3.1.2) as well. In brief, principal component analysis (PCA) of miRNA expression profile was performed by Factoextra package; heatmap analysis of miRNA expression profile was performed by pheatmap package; differentially expressed miRNAs were analyzed by Limma package, and miRNAs with a fold change (FC) ≥1.5 and an adjusted P value (FDR, False discovery rate) <0.1 were identified as differentially expressed miRNAs and exhibited by Volcano Plots according to the definition of previous studies (14,15); heatmap plot of differentially expressed miRNAs was performed by pheatmap package; enrichment analysis of dysregulated miRNAs was performed by Fisher exact test based on annotations from Gene Ontology (GO), Kyoko Encyclopedia of Genes and Genomes (KEGG), human-phenotype-ontology (HP), Disease Ontology (DOID).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) validation
Using the screening criteria: absolute value of FC >2 and the adjusted P (Padj) value <0.01, a total of 5 candidate miRNAs were selected from differentially expressed miRNAs in microarray, then those 5 candidate miRNAs were further verified in the total 60 RI-MM patients and 60 non-RI-MM patients by RT-qPCR. In brief, total RNA was extracted from plasma cells using TRIzol™ Reagent (Thermo Fisher Scientific, Waltham, USA), and then cDNA was reversely transcribed by QuantiTect Rev. Transcription Kit (Qiagen, Frankfurt, Germany). Subsequently, polymerase chain reaction was performed using Direct SYBR® Premix (Clontech, Mountain View, USA). All the procedures were in line with the instructions of the manufacturers. The primers used in RT-qPCR were listed in the Table 1. The miRNA expression was then calculated by 2−11Ct with the U6 as the internal reference.
Table 1
| Gene | Forward (5'→3') | Reverse (5'→3') |
|---|---|---|
| hsa-miR-103a-3p | ACACTCCAGCTGGGAGCAGCATTGTACAGG | TGTCGTGGAGTCGGCAATTC |
| hsa-miR-449c-5p | ACACTCCAGCTGGGTAGGCAGTGTATTGCT | TGTCGTGGAGTCGGCAATTC |
| hsa-miR-877-5p | ACACTCCAGCTGGGGTAGAGGAGATGGCGC | TGTCGTGGAGTCGGCAATTC |
| hsa-miR-455-3p | ACACTCCAGCTGGGGCAGTCCATGGGCATA | TGTCGTGGAGTCGGCAATTC |
| hsa-let-7a-5p | ACACTCCAGCTGGGTGAGGTAGTAGGTTGT | TGTCGTGGAGTCGGCAATTC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
miRNA, microRNA.
Statistical analysis
Kolmogorov-Smirnov test was performed to determine the normality of quantitative data. Normally distributed data were expressed as mean and standard deviation (SD), while the non-normally distributed data were displayed as median and inter-quartile range (IQR). And the qualitative data were presented as number (percentage). The student’s t test and Wilcoxon rank sum test were used to determine the comparison of quantitative data between two groups as appropriate, and the Chi-square test was applied to determine the comparison of qualitative data between two groups. Receiver operating characteristic (ROC) curve and the derived area under the curve (AUC) were used to assess the predicting ability of miRNAs for RI risk in MM patients. SPSS 21.0 statistical software (IBM, New York, USA) was used for statistical data processing, and the GraphPad Prism 7.02 (GraphPad Software Inc., New York, USA) was applied for plotting graphs. All tests were two-sided, and P value <0.05 was considered as significant.
Results
Patients’ characteristics in microarray assay
A total of 20 RI-MM patients and 20 non-RI-MM patients were included in the microarray assay as the detailed characteristics shown in Table 2. In brief, no difference of age (P=0.806), gender (P=0.736), body weight (P=0.643), calcium (P=0.314) was observed between RI-MM patients and non-RI-MM patients, while Hb (P=0.004), ALB (P=0.005) and Ccr (P<0.001) were lower, Scr (P<0.001), β2-MG (P<0.001) and LDH (P=0.004) were higher in RI-MM patients compared to non-RI-MM patients.
Table 2
| Items | Microarray assay | RT-qPCR validation | |||||
|---|---|---|---|---|---|---|---|
| RI-MM patients (N=20) | Non-RI-MM patients (N=20) | P value | RI-MM patients (N=60) | Non-RI-MM patients (N=60) | P value | ||
| Age (years), mean ± SD | 56.6±7.4 | 57.3±9.1 | 0.806 | 57.7±7.4 | 55.8±8.9 | 0.222 | |
| Gender, No. (%) | 0.736 | 0.336 | |||||
| Female | 7 (35.0) | 6 (30.0) | 23 (38.3) | 18 (30.0) | |||
| Male | 13 (65.0) | 14 (70.0) | 37 (61.7) | 42 (70.0) | |||
| Body weight (kg), mean ± SD | 60.9±9.5 | 62.4±11.6 | 0.643 | 59.0±10.5 | 60.3±10.4 | 0.486 | |
| Hb (g/dL), mean ± SD | 8.8±2.3 | 10.8±1.8 | 0.004 | 9.8±2.4 | 10.4±2.5 | 0.168 | |
| Calcium (mg/dL), mean ± SD | 9.9±1.6 | 10.4±1.3 | 0.314 | 10.1±1.7 | 10.1±1.9 | 0.962 | |
| Scr (mg/dL), median (IQR) | 2.8 (2.4–3.0) | 1.4 (1.0–1.6) | <0.001 | 3.0 (2.5–3.6) | 1.4 (1.2–1.7) | <0.001 | |
| Ccr (mL/min), median (IQR) | 25.2 (22.1–30.8) | 49.0 (41.1–68.8) | <0.001 | 23.2 (17.8–26.7) | 45.6 (41.6–54.2) | <0.001 | |
| ALB (mg/dL), mean ± SD | 3.2±0.7 | 3.8±0.7 | 0.005 | 3.2±0.8 | 4.0±0.7 | <0.001 | |
| β2-MG (mg/L), median (IQR) | 5.6 (4.4–9.1) | 2.7 (1.1–3.8) | <0.001 | 6.3 (4.1–9.8) | 2.7 (1.2–4.4) | <0.001 | |
| LDH (U/L), median (IQR) | 227.2 (190.0–285.9) | 182.9 (125.3–218.3) | 0.004 | 224.9 (194.3–258.7) | 176.5 (140.4–207.6) | <0.001 | |
Normality was determined by Kolmogorov-Smirnov test. Comparison was determined by Student’s t test, Chi-square test or Wilcoxon rank sum test. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MM, multiple myeloma; RI, renal impairment; SD, standard deviation; Hb, hemoglobin; IQR, interquartile range; Scr, serum creatinine; Ccr, creatinine clearance; ALB, albumin; β2-MG, beta‐2‐microglobulin; LDH, lactate dehydrogenase.
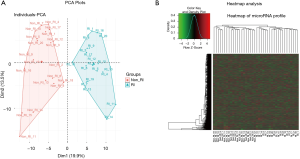
PCA and heatmap analysis of miRNA expression profile
PCA plots exhibited that miRNA expression profile could clearly distinguish RI-MM patients from non-RI-MM patients (Figure 1A), and heatmap analysis also showed that miRNA expression profile could obviously differentiate RI-MM patients from non-RI-MM patients (Figure 1B).
Valcano plots and heatmap analysis of differentially expressed miRNAs
Valcano plots identified 28 upregulated and 13 downregulated miRNAs in RI-MM patients compared to non-RI-MM patients (Figure 2A), and heatmap illuminated that these differentially expressed miRNAs clearly differentiate RI-MM patients from non-RI-MM patients (Figure 2B). Besides, the detailed information of these differentially expressed miRNAs were shown in Table 3.
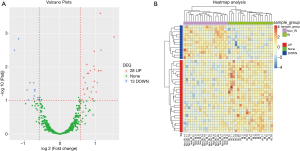
Table 3
| miRNA name | Probe ID | Log2FC | P value | Adjusted P value | Trend |
|---|---|---|---|---|---|
| hsa-miR-103a-3p | MIMAT0000101 | 1.14348 | 4.84E-07 | 0.00025 | UP |
| hsa-miR-449c-5p | MIMAT0010251 | 1.533664 | 2.94E-06 | 0.001263 | UP |
| hsa-miR-877-5p | MIMAT0004949 | −1.16843 | 3.91E-06 | 0.00144 | DOWN |
| hsa-miR-455-3p | MIMAT0004784 | −1.28229 | 1.10E-05 | 0.003157 | DOWN |
| hsa-let-7a-5p | MIMAT0000062 | 1.165801 | 1.10E-05 | 0.003157 | UP |
| hsa-miR-151a-3p | MIMAT0000757 | 0.972224 | 1.30E-05 | 0.003364 | UP |
| hsa-miR-20b-5p | MIMAT0001413 | 0.760544 | 3.59E-05 | 0.008425 | UP |
| hsa-miR-320a | MIMAT0000510 | 0.962337 | 4.39E-05 | 0.009424 | UP |
| hsa-miR-620 | MIMAT0003289 | 0.979332 | 6.01E-05 | 0.01191 | UP |
| hsa-miR-488-5p | MIMAT0002804 | 0.865665 | 7.62E-05 | 0.012839 | UP |
| hsa-miR-17-5p | MIMAT0000070 | 1.09423 | 7.23E-05 | 0.012839 | UP |
| hsa-miR-663a | MIMAT0003326 | 1.158985 | 7.97E-05 | 0.012839 | UP |
| hsa-let-7g-5p | MIMAT0000414 | 1.089788 | 8.54E-05 | 0.012955 | UP |
| hsa-miR-559 | MIMAT0003223 | 0.77763 | 0.000114 | 0.015509 | UP |
| hsa-miR-127-3p | MIMAT0000446 | 0.809156 | 0.000114 | 0.015509 | UP |
| hsa-miR-146a-5p | MIMAT0000449 | 0.672637 | 0.000159 | 0.020467 | UP |
| hsa-miR-520a-3p | MIMAT0002834 | 0.974852 | 0.000216 | 0.026478 | UP |
| hsa-miR-155-5p | MIMAT0000646 | −0.84853 | 0.000276 | 0.029466 | DOWN |
| hsa-miR-4463 | MIMAT0018987 | −0.73943 | 0.000286 | 0.029466 | DOWN |
| hsa-miR-4640-5p | MIMAT0019699 | 0.720609 | 0.000273 | 0.029466 | UP |
| hsa-miR-3128 | MIMAT0014991 | 1.059139 | 0.000267 | 0.029466 | UP |
| hsa-miR-181a-2-3p | MIMAT0004558 | 0.9362 | 0.000401 | 0.03978 | UP |
| hsa-miR-361-3p | MIMAT0004682 | 0.783039 | 0.000442 | 0.042082 | UP |
| hsa-miR-190a-3p | MIMAT0026482 | 0.667474 | 0.000532 | 0.045674 | UP |
| hsa-miR-520b | MIMAT0002843 | 0.769241 | 0.000514 | 0.045674 | UP |
| hsa-miR-6865-5p | MIMAT0027630 | −0.71012 | 0.000562 | 0.046769 | DOWN |
| hsa-miR-642a-3p | MIMAT0020924 | −0.92327 | 0.000597 | 0.048087 | DOWN |
| hsa-miR-6511a-5p | MIMAT0025478 | −0.70514 | 0.000665 | 0.051972 | DOWN |
| hsa-miR-4423-3p | MIMAT0018936 | 0.885864 | 0.000745 | 0.054903 | UP |
| hsa-miR-6799-3p | MIMAT0027499 | 1.024948 | 0.000744 | 0.054903 | UP |
| hsa-miR-326 | MIMAT0000756 | 0.694285 | 0.000903 | 0.064696 | UP |
| hsa-miR-7154-5p | MIMAT0028218 | −0.72389 | 0.001046 | 0.072906 | DOWN |
| hsa-miR-92a-3p | MIMAT0000092 | 0.833052 | 0.001207 | 0.079787 | UP |
| hsa-miR-6511a-3p | MIMAT0025479 | −0.62136 | 0.001289 | 0.081048 | DOWN |
| hsa-miR-1268a | MIMAT0005922 | −0.74005 | 0.00145 | 0.087705 | DOWN |
| hsa-miR-181c-5p | MIMAT0000258 | 0.933365 | 0.001463 | 0.087705 | UP |
| hsa-miR-1281 | MIMAT0005939 | 0.917223 | 0.00155 | 0.090821 | UP |
| hsa-miR-4775 | MIMAT0019931 | −0.62303 | 0.00166 | 0.092911 | DOWN |
| hsa-miR-508-5p | MIMAT0004778 | −0.58759 | 0.001799 | 0.094644 | DOWN |
| hsa-miR-96-5p | MIMAT0000095 | 0.623519 | 0.001837 | 0.094705 | UP |
| hsa-miR-6511b-5p | MIMAT0025847 | −0.88698 | 0.001882 | 0.095124 | DOWN |
miRNA, microRNA; ID, identity; FC, fold change.
Enrichment analysis
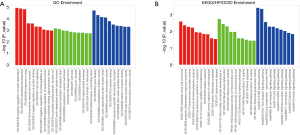
GO enrichment analysis illuminated that the differentially expressed miRNAs were enriched in molecular function (such as cyclin binding, endopeptidase inhibitor activity, cyclin dependent protein kinase inhibitor activity, etc.), cellular component (such as cyclin dependent protein kinase holoenzyme complex, eukaryotic translation elongation factor 1 complex, cytoplasmic vesicle membrane, etc.) and biological process (such as negative regulation of cyclin dependent protein kinase activity, osteoblast development, negative regulation of apoptotic process, etc.) (Figure 3A). KEGG enrichment analysis illustrated that the differentially expressed miRNAs were enriched in renal/inflammatory/apoptosis pathways such as ErbB signaling pathway, RIG I like receptor signaling pathway, B cell receptor signaling pathway, p53 signaling pathway, NOD like receptor signaling pathway, etc. (Figure 3B). HP enrichment analysis disclosed that the differentially expressed miRNAs were enriched in renal-dysfunction/mitochondrial related phenotypes such as renal tubular dysfunction, decreased activity of mitochondrial complex I, renal dysplasia, etc. (Figure 3B). Finally, DOID enrichment analysis revealed that the differentially expressed miRNAs were enriched in renal or inflammatory diseases such as metastatic renal cell carcinoma, systemic inflammatory response syndrome, autoimmune hemolytic anemia, immune thrombocytopenic purpura, etc. (Figure 3B).
Patients’ characteristics in RT-qPCR validation
A total of 60 RI-MM patients and 60 non-RI-MM patients were included in the RT-qPCR validation as the detailed characteristics shown in Table 2. In brief, no difference of age (P=0.222), gender (P=0.336), body weight (P=0.486), Hb (P=0.168), calcium (P=0.962) was observed between RI-MM patients and non-RI-MM patients, while ALB (P<0.001) and Ccr (P<0.001) were lower, Scr (P<0.001), β2-MG (P<0.001) and LDH (P<0.001) were higher in RI-MM patients compared to non-RI-MM patients.
Expressions of candidate miRNAs
Using the screening criteria: absolute value of FC >2 and the Padj value <0.01, a total of 5 candidate miRNAs were selected from differentially expressed miRNAs in microarray, then those 5 candidate miRNAs were further verified in the total 60 RI-MM patients and 60 non-RI-MM patients by RT-qPCR. As shown in Figure 4, miR-103a-3p, miR-449c-5p and let-7a-5p were greatly increased, while miR-877-5p and miR-455-3p were dramatically decreased in RI-MM patients compared to non-RI-MM patients (all P<0.001, Padj<0.001).
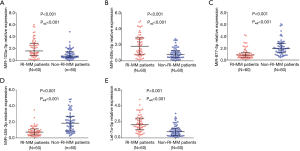
Predictive value of candidate miRNAs for RI risk in MM patients
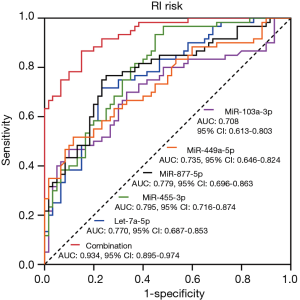
ROC curve analysis showed that miR-103a-3p, miR-449c-5p, miR-877-5p, miR-455-3p and let-7a-5p could all predict RI risk in MM patients, most importantly, the combination of these five miRNAs presented with a great predictive value for RI risk in MM patients with an AUC of 0.934, 95% CI: 0.895–0.974 (Figure 5).
Discussion
In this study, we discovered that: (I) the expression profile of miRNA notably differentiated RI-MM patients from non-RI MM patients; (II) 28 upregulated and 13 downregulated miRNAs in RI-MM patients compared to non-RI-MM patients were identified, which were mainly enriched in renal/inflammatory/apoptosis pathways and renal/inflammatory diseases; (III) furthermore, 5 differentially expressed miRNAs in microarray were selected as candidate miRNAs for RT-qPCR validation in a large sample size population, which displayed that there were 3 upregulated miRNAs (miR-103a-3p, miR-449c-5p, let-7a-5p) and 2 downregulated miRNAs (miR-877-5p and miR-455-3p) in RI-MM patients compared with non-RI MM patients; then the ROC curve analysis disclosed that combining the 5 candidate miRNAs had great value for predicting the RI risk in MM patients.
RI is a frequent comorbidity and complication in MM as a result of the insufficient renal function derived from the accumulation of immunoglobulin light chains in the kidney, which often results in the cast nephropathy (5,16). It has been reported that RI attacks approximately 20–50% MM patients at diagnosis and is correlated with unsatisfactory clinical outcome and survival profiles, especially for the patients who are already complicated with some chronic disease, which could lead to a chronic damage in the renal function (2,3,17-19). Furthermore, treatment of RI in MM is associated with better survival in patients, for instance, a recent clinical trial reveals that pomalidomide accompanied with low-dose dexamethasone achieves good efficacy in regard to treatment response and overall survival (20). Thus, an early identification of RI in MM patients is necessary in enhancing clinical outcome and prolonging survival of MM patients.
To better improve diagnosis and treatment in RI-MM patients, increasing studies investigating the potential biomarkers for predicting RI in MM patients, however, the predictive value of miRNA for RI in MM patients is still unclear. Nonetheless, there are several studies which report the miRNA expression profile in RI previously. For instance, a recent study identifies 9 miRNAs that are dysregulated among diabetic nephropathy (DN) patients, diabetic patients with membranous nephropathy and patients with normal histology, and 2 of these miRNAs are validated to be involved in the regulation of kidney fibrosis by interacting with ubiquitin-conjugating E2 enzyme variant (UBE2v1) (21). Another study that investigates the miRNA expression profile in the rat models with acute kidney injury identifies 22 downregulated miRNAs and 19 upregulated miRNAs in kidney (22). A study using TaqMan low-density array (TLDA) shows that there are 11 upregulated miRNAs and 11 downregulated miRNAs in plasma of patients with sepsis-induced kidney injury compared with healthy controls (23). In the present study, we investigated the miRNA expression profile in RI-MM patients and found the miRNA expression profile markedly differentiated RI-MM patients from non-RI MM patients, and the dysregulated miRNAs are enriched in renal/inflammatory/apoptosis pathways as well as renal/inflammatory diseases. These results indicate that miRNAs expression profile is markedly dysregulated in RI-MM patients, and may be critical regulators in the pathogenesis of RI in MM patients.
In order to further explore the value of differentially expressed miRNAs in RI-MM patients, 5 differentially expressed miRNAs in microarray were selected for RT-qPCR validation, which disclosed 3 upregulated miRNAs (miR-103a-3p, miR-449c-5p, let-7a-5p) and 2 downregulated miRNAs (miR-877-5p and miR-455-3p) in RI-MM patients compared with non-RI MM patients. Furthermore, the subsequent analyses elucidated that all 5 candidate miRNAs could differentiate RI-MM patients from non-RI MM patients, and their combination exhibited an excellent value for predicting the RI risk in MM patients. And here are several possible explanations to these results: (I) miR-103a-3p: miR-103a has been previously illuminated as a factor promoting inflammation and fibrosis in kidney. For instance, a prior experiment shows that circulating miR-103a expression could be upregulated by angiotensin II, which subsequently results in increased SNRK level in glomerular endothelial cells, leading to renal inflammation and fibrosis in mouse models (24); (II) miR-449c-5p: it has been reported that somatostatin receptor subtype 5 (SSTR5) could inhibit the CRH receptor subtype 1 (CRHR1) expression and function by inducing miR-449c, which subsequently advocates the secondary adrenal dysfunction (25); (III) let-7a-5p: let-7a has been found to be a regulator promoting renal dysfunction. For example, an experiment demonstrates that naringenin reduces kidney injury by regulating the let-7a/ transforming growth factor-β1 receptor 1 (TGFBR1) signaling pathway in diabetic nephropathy rats (26). And another experiment reveals that let-7a enhances hyperplasia and inflammation by increasing cell proliferation and NF-kB activation in systemic lupus erythematosus cell models (27); (IV) miR-877-3p: previous studies report that miR-877 may play an essential role in preventing the progression of RI. For instance, miR-877-3p downregulation results in the over production of interleukin (IL)-1β in human mesangial cells (obtained from IgA nephropathy patients) that are activated by secretory IgA (28). And another experiment reveals that miR-877 inhibits renal cell cancer cell proliferation and migration via mediating the eukaryotic elongation factor-2 kinase (eEF2K) /eEF2 signaling pathway (29); (V) miR-445-3p: miR-455-3p is reported to inhibit renal fibrosis via suppressing the Rho-associated coiled coil-containing protein kinase 2 (ROCK2) expression in diabetic nephropathy rats (30). Additionally, miR-445-3p and miR-445-5p are illuminated to suppress cancer cell proliferation and invasive abilities through targeting SKA1 and SKA3 in renal cell carcinoma (31).
Besides, there existed several limitations in this study: (I) the statistical power might be slightly diminished due the relatively small sample of our study; (II) the precise molecular functions of the 5 candidate miRNAs including miR-103a-3p, miR-449c-3p, let-7a-5p, miR-877-5p and miR445-3p in the development or progression of RI in MM were not evaluated in this study; (III) the prognostic roles of the 5 candidate miRNAs for RI-MM patients were not analyzed in our study.
In conclusion, miRNA expression profile is closely implicated in the RI development, and miR-103a-3p, miR-449c-5p, miR-877-5p, miR-455-3p as well as let-7a-5p may serve as novel biomarkers for RI risk in MM patients.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.41). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Institutional Review Board of our hospital with ethical approval number 2015-018, and all patients provided the written informed consents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Röllig C, Knop S, Bornhauser M. Multiple myeloma. Lancet 2015;385:2197-208. [Crossref] [PubMed]
- Fotiou D, Dimopoulos MA, Kastritis E. Managing renal complications in multiple myeloma. Expert Rev Hematol 2016;9:839-50. [Crossref] [PubMed]
- Shi H, Chen Z, Xie J, et al. The Prevalence and Management of Multiple Myeloma-Induced Kidney Disease in China. Kidney Dis (Basel) 2016;1:235-40. [Crossref] [PubMed]
- Yadav P, Cook M, Cockwell P. Current Trends of Renal Impairment in Multiple Myeloma. Kidney Dis (Basel) 2016;1:241-57. [Crossref] [PubMed]
- Gavriatopoulou M, Terpos E, Kastritis E, et al. Current treatments for renal failure due to multiple myeloma. Expert Opin Pharmacother 2016;17:2165-77. [Crossref] [PubMed]
- Gholaminejad A, Abdul Tehrani H, Gholami Fesharaki M. Identification of candidate microRNA biomarkers in renal fibrosis: a meta-analysis of profiling studies. Biomarkers 2018;23:713-24. [Crossref] [PubMed]
- Petrica L, Pusztai AM, Vlad M, et al. MiRNA Expression is Associated with Clinical Variables Related to Vascular Remodeling in the Kidney and the Brain in Type 2 Diabetes Mellitus Patients. Endocr Res 2019. [Epub ahead of print].
- Jaurretche S, Perez G, Antongiovanni N, et al. Variables Associated with a Urinary MicroRNAs Excretion Profile Indicative of Renal Fibrosis in Fabry Disease Patients. Int J Chronic Dis 2019;2019:4027606.
- Liu H, Wang G, Huang Y, et al. Identification specific miRNA in t(4;14) multiple myeloma based on miRNA-mRNA expressing profile correlation analysis. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]
- Musolino C, Oteri G, Allegra A, et al. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann Hematol 2018;97:1259-69. [Crossref] [PubMed]
- Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol 2014;15:e538-48. [Crossref] [PubMed]
- Muñoz-Culla M, Irizar H, Saenz-Cuesta M, et al. SncRNA (microRNA &snoRNA) opposite expression pattern found in multiple sclerosis relapse and remission is sex dependent. Sci Rep 2016;6:20126. [Crossref] [PubMed]
- Hubal MJ, Nadler EP, Ferrante SC, et al. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity (Silver Spring) 2017;25:102-10. [Crossref] [PubMed]
- Ioannidis J, Donadeu FX. Circulating miRNA signatures of early pregnancy in cattle. BMC Genomics 2016;17:184. [Crossref] [PubMed]
- Ackerman WE, Buhimschi IA, Eidem HR, et al. Comprehensive RNA profiling of villous trophoblast and decidua basalis in pregnancies complicated by preterm birth following intra-amniotic infection. Placenta 2016;44:23-33. [Crossref] [PubMed]
- Acquah ME, Hsing AW, McGuire V, et al. Presentation and survival of multiple myeloma patients in Ghana: a review of 169 cases. Ghana Med J 2019;53:52-58. [Crossref] [PubMed]
- Graziani G, Herget GW, Ihorst G, et al. Time from first symptom onset to the final diagnosis of multiple myeloma (MM) - possible risks and future solutions: retrospective and prospective 'Deutsche Studiengruppe MM' (DSMM) and 'European Myeloma Network' (EMN) analysis. Leuk Lymphoma 2020;61:875-86. [PubMed]
- Courant M, Orazio S, Monnereau A, et al. Incidence, prognostic impact and clinical outcomes of renal impairment in patients with multiple myeloma: a population-based registry. Nephrol Dial Transplant 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Chen J, Liu H, Li L, et al. Clinical features and treatment outcome of elderly multiple myeloma patients with impaired renal function. J Clin Lab Anal 2019;33:e22888. [Crossref] [PubMed]
- Dimopoulos M, Weisel K, van de Donk N, et al. Pomalidomide Plus Low-Dose Dexamethasone in Patients With Relapsed/Refractory Multiple Myeloma and Renal Impairment: Results From a Phase II Trial. J Clin Oncol 2018;36:2035-43. [Crossref] [PubMed]
- Conserva F, Barozzino M, Pesce F, et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of Kidney Fibrosis in Diabetic Nephropathy. Sci Rep 2019;9:11357. [Crossref] [PubMed]
- Liu Y, Liu B, Liu Y, et al. MicroRNA expression profile by next-generation sequencing in a novel rat model of contrast-induced acute kidney injury. Ann Transl Med 2019;7:178. [Crossref] [PubMed]
- Lin Y, Ding Y, Song S, et al. Expression patterns and prognostic value of miR-210, miR-494, and miR-205 in middle-aged and old patients with sepsis-induced acute kidney injury. Bosn J Basic Med Sci 2019;19:249-56. [PubMed]
- Lu Q, Ma Z, Ding Y, et al. Circulating miR-103a-3p contributes to angiotensin II-induced renal inflammation and fibrosis via a SNRK/NF-kappaB/p65 regulatory axis. Nat Commun 2019;10:2145. [Crossref] [PubMed]
- Yamamoto M, Ben-Shlomo A, Kameda H, et al. Somatostatin receptor subtype 5 modifies hypothalamic-pituitary-adrenal axis stress function. JCI Insight 2018;3: [Crossref] [PubMed]
- Yan N, Wen L, Peng R, et al. Naringenin Ameliorated Kidney Injury through Let-7a/TGFBR1 Signaling in Diabetic Nephropathy. J Diabetes Res 2016;2016:8738760.
- Venugopal P, Koshy T, Lavu V, et al. Differential expression of microRNAs let-7a, miR-125b, miR-100, and miR-21 and interaction with NF-kB pathway genes in periodontitis pathogenesis. J Cell Physiol 2018;233:5877-84. [Crossref] [PubMed]
- Liang Y, Zhao G, Tang L, et al. MiR-100-3p and miR-877-3p regulate overproduction of IL-8 and IL-1beta in mesangial cells activated by secretory IgA from IgA nephropathy patients. Exp Cell Res 2016;347:312-21. [Crossref] [PubMed]
- Shi Q, Xu X, Liu Q, et al. MicroRNA-877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncol Lett 2016;11:1474-80. [Crossref] [PubMed]
- Wu J, Liu J, Ding Y, et al. MiR-455-3p suppresses renal fibrosis through repression of ROCK2 expression in diabetic nephropathy. Biochem Biophys Res Commun 2018;503:977-83. [Crossref] [PubMed]
- Yamada Y, Arai T, Kojima S, et al. Anti-tumor roles of both strands of the miR-455 duplex: their targets SKA1 and SKA3 are involved in the pathogenesis of renal cell carcinoma. Oncotarget 2018;9:26638-58. [Crossref] [PubMed]