
Overexpression of lncRNA H19 leads to reduced proliferation in TSCC cells through miR-675-5p/GPR55
Introduction
Tongue squamous cell carcinoma (TSCC) is a highly malignant tumor and oral disease. There are many different theories as to why and how TSCC occurs. These theories include environment factors, genetic factors and immune escape factors. Recently, long noncoding RNAs (lncRNAs) were proven to affect cell proliferation, invasion, migration, apoptosis and the cell cycle (1,2). LncRNA H19 exhibits abnormal expression in laryngeal squamous cell carcinoma (3), which is similar in location and type to squamous cell carcinoma of the tongue. Recent studies have also shown that lncRNA H19 is closely related to the occurrence and development of oral squamous cell carcinoma (4-6). H19 is a nonprotein-coding gene, and its longest transcript, lncRNA H19, is thought to be negatively correlated with many different tumors, including prostate cancer (7), human pancreatic ductal adenocarcinoma (8) and breast cancer (9). It has also been reported that lncRNA H19 inhibits tumor cell proliferation (7). However, some studies have indicated that lncRNA H19 promotes tumor progression (10-12), while others reported that lncRNA H19 might induce resistance to 1,25(OH)2D3 in colon cancer cells (13). Whether H19 functions in TSCC and/or how it regulates the occurrence and development of TSCC is unknown. The literature contends that lncRNA H19 can transcribe miR-675-5p. Through an online prediction tool, we found that miR-675-5p might bind to GPR55, which has been suggested as a growth promoter for TSCC in the literature.
We hypothesized that expression of lncRNA H19 may influence TSCC through miR-675-5p and GPR55, and aimed to explore the potential involvement of lncRNA H19 in growth regulation of TSCC cell lines through its exogenous overexpression.
Methods
Clinical sample collection
From January 2013 to February 2017, TSCC samples and adjacent normal tissues (ANTs) from 22 patients were obtained from the Department of Oral and Maxillofacial Surgery, Stomatology Hospital of Guangzhou Medical University, Guangdong Provincial Stomatological Hospital, Guangzhou First Peoples’ Hospital and the No. 5 Affiliated Hospital of Sun Yat-sen University. Patients had not previously received radiotherapy, chemotherapy, or any other treatment prior to surgery. Tumor tissue samples and ANTs (at least 2.0 cm distal to the tumor margin) were snap-frozen in liquid nitrogen for quantitative real time PCR (qRT-PCR) assays.
This study was approved by the Institutional Ethics Committee of the Stomatology Hospital of Guangzhou Medical University. The study was performed in accordance with the Declaration of Helsinki.
Cell culture
CAL27 and SCC9 cell lines were a gift from Professor Jinsong Li in the Department of Oral and Maxillofacial Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University. CAL27 and SCC9 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) at 37 °C and 5% CO2.
Design of the lncRNA H19 plasmid
LncRNA H19 (Gene ID: 283120 from NCBI) was amplified using gene-specific forward 5' CGGAATTCGCCACCGGGAGGGGGT 3' and reverse 5' GATTCTAGA GCTGTACAGTGTTTATTGATGATGA 3' primers (1 µM each). PCR products were verified using 1% agarose electrophoresis. The pcDNA3.1 (+) vector, lncRNA H19 and a nonsense sequence were all digested with EcoR I and XbaI. Then, they were joined together to construct a lncRNA H19 plasmid and a negative control plasmid (The plasmid was constructed by a company called Sagene, CHN). Resulting plasmids were then confirmed by genetic sequencing.
Overexpression of lncRNA H19
To confirm that lncRNA H19 functions as a tumor suppressor in TSCC, our team transfected CAL27 and SCC9 cells with the lncRNA H19 plasmid to overexpress H19 (lncRNA H19 group). Cells (1×105 cells per well) were grown to 90% confluence in 24-well plates and the transfected with lncRNA H19 (lncRNA H19 group) using Lipofectamine 2000 (Life Technologies Corporation, USA) according to the manufacturer’s instructions. As a negative control (negative control group), an additional group of cells was transfected with the negative control plasmid and the same amount of Lipofectamine 2000, and cells treated with only Lipofectamine were treated as the blank group. Culture cells were harvested in triplicate after 24, 48, and 72 h of transfection.
RNA extraction and quantitative real-time PCR
Freshly frozen clinical samples were ground, and total RNA was extracted using TRIzol reagent (Invitrogen, USA). CAL27 and SCC9 cells were seeded into six-well plates at a density of 5×104 cells/well for quantitative real time PCR (qRT-PCR) assays. Cells overexpressing lncRNA H19 were harvested at 80% confluence, and RNA was extracted as described above. First-strand cDNA was synthesized using the GeneAmp® PCR System 9700 (Life Technologies Corporation, USA). Primers used for the reaction were as follows: LncRNA H19 (forward: 5'-CGGAATTCGCCACCGGGAGGGGGT -3' and reverse: 5'-GATTCTAGA GCTGTACAGTGTTTATTGATGATGA-3'); miR-675-5p (forward: 5'-ACACTCCAGCTGGGCTGTATGCCCTCAC-3' and reverse 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGAGCGGT-3'); and β-actin (forward: 5'-CCATCGTCCACCGCAAAT-3' and reverse (5'-CCTGTAACAACGCATCTCATA-3'). Quantitative real-time PCR was performed using StepOne and a StepOnePlus Real-Time PCR system (Life Technologies Corporation, USA). Finally, expression levels of lncRNA H19 and miR-675-5p were calculated using the 2-∆∆Ct method. Each experiment was repeated at least three times.
MTT assay
The two cell lines were transfected with lncRNA H19 (lncRNA H19 group), the negative control plasmid (negative control group) or transfection reagents alone (blank group). Transfected cells were grown in 96-well plates, and cell proliferation was measured every 24 h using the Cell Proliferation Reagent Kit I (MTT, Roche, USA) according to the manufacturer’s instructions. The relative numbers of viable cells were calculated using the absorbance values (optical density) at 450 nm using an Easy Reader 340 AT (SLT-Lab Instruments). All experiments were performed in quadruplicate.
Transwell migration assay
To detect changes in the migratory ability of cells, the two cell lines were transfected with lncRNA H19 (lncRNA H19 group), the negative control plasmid (negative control group) or transfection reagents alone (blank group). Cells were harvested when they reached 80% confluence.
Cells were seeded at a density of 1×105 cells/well in the upper Transwell chambers. Then, upper Transwell chambers were placed onto the lower Transwell chambers containing 20% FBS. Twenty-four hours after seeding, cells that had migrated to the lower Transwell chamber were counted using crystal violet staining solution (BD Biosciences, USA).
Transwell invasion assay
To determine changes in the invasion ability of cells, the two cell lines were transfected with lncRNA H19 (lncRNA H19 group), the negative control plasmid (negative control group) or transfection reagents alone (blank group). Cells were harvested when they reached 80% confluence. Before seeding the cells, Matrigel (BD Biosciences, USA) was placed in the bottom of the upper Transwell chamber to simulate the in vivo extracellular matrix (ECM). Subsequent steps were similar to the Transwell migration assay described above.
Dual-luciferase reporter gene assay
Luciferase activity was measured using a Dual-Luciferase Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions to investigate the existence of binding sites between miR-675-5p and GPR55.
Statistical analysis
Differences between the three groups were compared using one-way ANOVA. GraphPad Prism (version 6.0, USA) was used to analyze the data and to construct the graphs. P values <0.05 were considered statistically significant.
Results
LncRNA H19 is expressed at low levels in TSCC
We evaluated lncRNA H19 expression in 22 paired TSCC tissues and ANTs and found that H19 was expressed at significantly lower levels in TSCC tissues compared to ANTs (Figure 1, P<0.01).
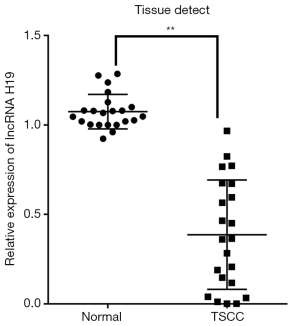
Plasmid construction
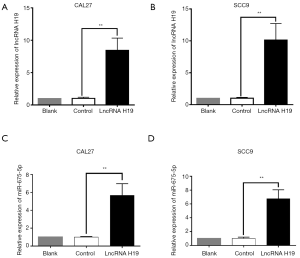
Agarose electrophoresis analyses showed that lncRNA H19 (2.3 kb) was successfully incorporated into the pcDNA3.1 (+) vector. In addition, genetic sequencing of the vector showed that the sequence was precisely that of lncRNA H19. After transfecting the plasmid, expression levels of lncRNA H19 were upregulated in both CAL27 and SCC9 cell lines (Figure 2A,C, P<0.05). Together, these data showed that the plasmid overexpressing lncRNA H19 was successfully constructed. In addition, expression levels of miR-675-5p were upregulated in both CAL27 and SCC9 cell lines (Figure 2B,D, P<0.05). These results are consistent with the hypothesis of Cai (14) that lncRNA H19 is a primary microRNA precursor of miR-675-5p, which is now well known; this indicates that when lncRNA H19 is upregulated, expression of miR-675-5p is also upregulated.
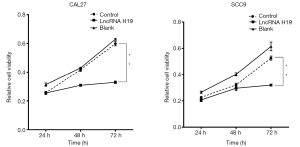
Upregulation of lncRNA H19 expression inhibits TSCC cell proliferation
MTT assay results showed that cell proliferation of CAL27 and SCC9 cell lines was reduced 72 h after lncRNA H19 overexpression (Figure 3A,B, lncRNA H19 group), while in the blank and negative control groups, cell proliferation of CAL27 and SCC9 cell lines was not affected (Figure 3A,B, blank group and negative control group). Differences between the three groups were significant (P<0.05). Therefore, we proposed that lncRNA H19 is negatively correlated with TSCC progression.
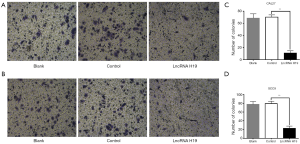
Upregulation of lncRNA H19 expression inhibits TSCC cell migration
After 24 h, cells below the Transwell chamber were counted. The number of migrated cells was significantly lower in the lncRNA H19-overexpressing groups than in the blank and negative control groups (Figure 4A,C). The specific number of migrated cells for each group is represented by corresponding histograms (Figure 4B,D). The results of Transwell assays demonstrated that cell migration ability was inhibited in response to lncRNA H19 overexpression.

Upregulation of lncRNA H19 expression inhibits TSCC cell invasion
After 24 h, the number of cells that invaded the Matrigel (BD Biosciences, USA) was significantly lower in the lncRNA H19-overexpressing group than in the blank and negative control groups (Figure 5A,C). The specific number of each group are represented by corresponding histograms (Figure 5B,D). Transwell assay results showed that cell invasion was inhibited in response to lncRNA H19 overexpression.
GPR55 is a direct target of miR-675-5p
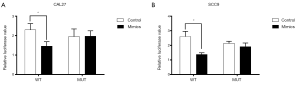
An online prediction tool showed that miR-675-5p and GPR55 may have several binding sites (http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000392040.1&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1). In addition, according to dual-luciferase reporter gene assays in CAL27 and SCC9 cells, luciferase activity was reduced in the GPR55-3'UTR-Wt+ miR-675-5p mimic group compared to the GPR55-3'UTR-Wt+ mimic NC group (Figure 6A,B, P<0.05). In addition, in the GPR55-3'UTR-Mut+ miR-675-5p mimic and GPR55-3'UTR-Mut+ mimic NC groups, luciferase activity was unchanged (Figure 6A,B). These data thus reveal that miR-675-5p binds to GPR55 and inhibits its expression.
Discussion
Long noncoding RNAs, which do not code for proteins, were ignored for many years due to their unknown function in cells (15). With the development of gene detection techniques, an increasing number of lncRNAs have been identified and proven to play a role in regulating gene expression at the epigenetic, transcriptional and posttranscriptional levels. These lncRNAs influence physiologic and pathologic events related to cellular growth, proliferation, differentiation, apoptosis and injury.
LncRNA H19 is reportedly negatively correlated with the occurrence and development of many different tumors (8,16-18). First, researchers confirmed that lncRNA H19 inhibited tumor growth in mice (19). In a prostate cancer study, researchers found that lncRNA H19 suppressed cancer metastasis by targeting TGFBI (7). Moreover, in gastric cancer, human pancreatic ductal adenocarcinoma, and breast cancer, lncRNA H19 was shown to mediate anti-tumor effects (8,9,20,21). However, no comprehensive study of the relationship between lncRNA H19 and TSCC progression exists. In this study, lncRNA H19 exhibited abnormal expression in TSCC (Figure 1), indicating that lncRNA H19 might be a potential influential factor in TSCC. In our initial experiments, we found that lncRNA H19 expression was reduced in TSCC tissues compared to normal tissues (Figure 1). Next, we observed that overexpression of lncRNA H19 inhibited proliferation in TSCC cell lines CAL27 and SCC9 (Figure 3). Therefore, we speculated that lncRNA H19 might also exert anti-tumor effects in TSCC.
TSCC is malignant not only due to its unlimited reproductive capacity but also due to its considerable effects on local invasion and cervical lymph node metastasis. Local invasion causes TSCC to develop quickly in only a few weeks or months in the tongue. Its migratory ability facilitates TSCC to metastasis to the cervical lymph nodes; for this reason, recurrence rates in TSCC patients are relatively high, even after successful resection (22). The number of cells that migrated through the Transwell chamber was decreased in response to lncRNA H19 overexpression (Figure 4). These results indicate that lncRNA H19 might inhibit migration in TSCC. In addition, results were similar for invasion: the number of cells that penetrated the Matrigel (BD Biosciences, USA) was significantly decreased in response to increased H19 expression (Figure 5).
Despite these findings, we were more interested in the mechanism through which lncRNA H19 influences TSCC development. Many studies have indicated that H19 plays a role in tumor regulation through miR-675-5p. Research has indicated that lncRNA H19 can be transcribed into miR-675-5p. MiR-675-5p is transcribed from the first exon of lncRNA H19 and functions in the posttranscriptional regulation of H19 (14)(14)(14)[14][14][14][12][14]. In a glioma study, researchers found that H19-derived miR-675-5p regulates cell proliferation and migration through CDK6, supporting the hypothesis that miR-675-5p functions in glioma progression (23). In addition, Zhu et al. (7) showed that the lncRNA H19/miR-675-5p axis inhibited prostate cancer metastasis through targeting TGFBI. Ma et al. (8) showed that lncRNA H19 derived miR-675 regulates cell proliferation by downregulating E2F-1. Furthermore, in bladder cancer, the lncRNA H19/miR-675-5p axis accelerates cell proliferation by regulating p53 activation (12). Zhang et al. (24) found that the lncRNA H19/miR-675 axis is involved in promoting cardiomyocyte apoptosis.
Based on results we obtained in this study, we aimed to determine the probable mechanism by which lncRNA H19 inhibits TSCC cell proliferation. After considering a detailed review of lncRNA, we propose that lncRNA functions in cells in multiple ways. First, it may affect the expression of downstream genes through transcriptional interference. Second, it may interfere with translation by hybridizing with its target mRNAs. Third, it may regulate protein activity by binding to special regions within the protein. Fourth, it may act as a precursor for various types of small RNAs (25,26). Zhou et al. (27) found that lncRNA H19 alters DNA methylation in the genome to change expression patterns of specific genes. Xia et al. (28) found that lncRNA H19 may act as a competing endogenous RNA (ceRNA) in translation, potentially playing important roles in posttranscriptional regulation in gastric cancer. In addition, Giovarelli et al. (29) found that lncRNA H19 controls mRNA decay by promoting the function of the KH-type splicing regulatory protein (KSRP). Regarding interactions with proteins, Zou et al. (30) found that lncRNA H19 regulates intestinal epithelial barrier function by interacting with the RNA-binding protein HuR. Moreover, lncRNA H19 can act as a miRNA sponge (31), interacting with several types of miRNAs (32). Recently, Wang et al. (33) found that lncRNA H19 inhibits fetal liver cell proliferation through the Wnt/β-catenin signaling pathway.
MicroRNAs function primarily by inhibiting protein translation. Through an online prediction tool, we found that miR-675-5p and GPR55 may have several binding sites (http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000392040.1&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1). Dual-luciferase reporter gene assays convincingly demonstrated that miR-675-5p binds to GPR55 (Figure 6).
GPR55 belongs to the G protein-coupled receptor family. It has important functions in the signaling and activation of proinflammatory cells and influences the development of cancer. In studies on the regulation of cell proliferation, Pérez-Gómez et al. (34) found that GPR55 drives skin carcinogenesis and is upregulated in human squamous cell carcinoma. Some research has indicated that GPR55 promotes cell proliferation as well. Hu et al. (35) found that the putative cannabinoid receptor GPR55 promotes cancer cell proliferation, and Andradas et al. (36) found that GPR55 promotes cancer cell proliferation via ERK. In addition, Ferro et al. (37) found that GPR55 signaling promotes proliferation of pancreatic cancer cells and tumor growth in mice. Additionally, in studies on the regulation of cancer metastasis, Kargl et al. (38,39) found that GPR55 promotes migration and adhesion in colon cancer cells, and Zhou et al. (40) found that the LPI/GPR55 axis enhances human breast cancer cell migration. Collectively, these findings indicate that GPR55 functions in tumor metastasis. Andradas et al. (41) found that GPR55 promotes metastasis in triple-negative breast cancer. TSCC, which is a human squamous cell carcinoma, may share the same mechanism as GPR55 in promoting tumor development. In a study on small-cell lung cancer, He et al. (42) found that expression of miRNAs in cancer tissue was reduced compared to normal tissue, and further study found that miR-675-5p may promote consistent cancer growth by specifically binding to GPR55 and reducing expression of GPR55. In this study, when lncRNA H19 was upregulated, miR-675-5p expression was also upregulated. Therefore, miR-675-5p decreases expression levels of GPR55, potentially resulting in reduced proliferation, invasion and migration of TSCC cells.
Despite these findings, the occurrence, development and progression of tumors is a very complex and diverse process. Our current study on TSCC shows only that lncRNA H19 is negatively correlated with growth, migration and invasion of TSCC, suggesting a possible relationship between lncRNA H19 and TSCC. This study is entirely performed using in vitro cell experiments, and related research has not yet expanded to animal experiments. In cell culture experiments, we overexpressed lncRNA H19 but did not perform reverse silencing verification studies of lncRNA H19. In addition, detection of GPR55 stopped at the binding site of miR-675-5p, a product of lncRNA H19, without further exploration. The results of our tissue examination demonstrate abnormal expression of lncRNA H19 in TSCC but did not combine tumor staging and survival curves for patients, and the results were one-sided. In the future, additional studies are necessary to further explore the relationship between lncRNA H19/miR-675-5p/GPR55 and TSCC. We expect that our research will provide the reference and the basis for pharmacologic and genetic therapy of TSCC.
Acknowledgments
The cell lines were donated by Jinsong Li of the Department of Oral and Maxillofacial Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
Funding: This work was financially supported by
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.45). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Ethics Committee of the Stomatology Hospital of Guangzhou Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang X, Ho TT. Computational Analysis of lncRNA Function in Cancer. Methods Mol Biol 2019;1878:139-55. [Crossref] [PubMed]
- Koch L. Functional genomics: Screening for lncRNA function. Nat Rev Genet 2017;18:70. [PubMed]
- Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget 2016;7:11553-66. [PubMed]
- Yuan Z, Yu Y, Zhang B, et al. Genetic variants in lncRNA H19 are associated with the risk of oral squamous cell carcinoma in a Chinese population. Oncotarget 2018;9:23915-22. [Crossref] [PubMed]
- Kou N, Liu S, Li X, et al. H19 facilitates tongue squamous cell carcinoma migration and invasion via sponging miR-let-7. Oncol Res 2019;27:173-82. [Crossref] [PubMed]
- Zhang DM, Lin ZY, Yang ZH, et al. IncRNA H19 promotes tongue squamous cell carcinoma progression through β-catenin/GSK3β/EMT signaling via association with EZH2. Am J Transl Res 2017;9:3474-86. [PubMed]
- Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014;281:3766-75. [Crossref] [PubMed]
- Ma L, Tian X, Guo H, et al. Long noncoding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J Cancer 2018;9:389-99. [Crossref] [PubMed]
- Wang J, Wang X, Chen T, et al. Huaier Extract Inhibits Breast Cancer Progression Through a LncRNA-H19/MiR-675-5p Pathway. Cell Physiol Biochem 2017;44:581-93. [Crossref] [PubMed]
- Cui J, Mo J, Luo M, et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:12400-9. [PubMed]
- Matouk IJ, Raveh E, Abu-lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 2014;1843:1414-26. [Crossref] [PubMed]
- Liu C, Chen Z, Fang J, et al. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol 2016;37:263-70. [Crossref] [PubMed]
- Chen S, Bu D, Ma Y, et al. H19 Overexpression Induces Resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in Colon Cancer Cells. Neoplasia 2017;19:226-36. [Crossref] [PubMed]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007;13:313-6. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res 2000;28:1221-7. [Crossref] [PubMed]
- Li T, Mo X, Fu L, et al. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016;7:8601-12. [PubMed]
- Yoshimizu T, Miroglio A, Ripoche MA, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A 2008;105:12417-22. [Crossref] [PubMed]
- Gabory A, Ripoche MA, Le Digarcher A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009;136:3413-21. [Crossref] [PubMed]
- Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci U S A 2011;108:16759-64. [Crossref] [PubMed]
- Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol 2014;20:5411-9. [Crossref] [PubMed]
- Sun Y, Han J, Lu Y, et al. Biological characteristics of a cell subpopulation in tongue squamous cell carcinoma. Oral Dis 2012;18:169-77. [Crossref] [PubMed]
- Li C, Lei B, Huang S, et al. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res 2015;7:1747-64. [PubMed]
- Zhang Y, Zhang M, Xu W, et al. The long non-coding RNA H19 promotes cardiomyocyte apoptosis in dilated cardiomyopathy. Oncotarget 2017;8:28588-94. [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009;23:1494-504. [Crossref] [PubMed]
- Zhou J, Yang L, Zhong T, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun 2015;6:10221. [Crossref] [PubMed]
- Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 2014;4:6088. [Crossref] [PubMed]
- Giovarelli M, Bucci G, Ramos A, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A 2014;111:E5023-8. [Crossref] [PubMed]
- Zou T, Jaladanki SK, Liu L, et al. H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Mol Cell Biol 2016;36:1332-41. [Crossref] [PubMed]
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. [Crossref] [PubMed]
- Imig J, Brunschweiger A, Brummer A, et al. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat Chem Biol 2015;11:107-14. [Crossref] [PubMed]
- Wang S, Wu X, Liu Y, et al. Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway. FEBS Lett 2016;590:559-70. [Crossref] [PubMed]
- Pérez-Gómez E, Andradas C, Flores JM, et al. The orphan receptor GPR55 drives skin carcinogenesis and is upregulated in human squamous cell carcinomas. Oncogene 2013;32:2534-42. [Crossref] [PubMed]
- Hu G, Ren G, Shi Y. The putative cannabinoid receptor GPR55 promotes cancer cell proliferation. Oncogene 2011;30:139-41. [Crossref] [PubMed]
- Andradas C, Caffarel MM, Perez-Gomez E, et al. The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene 2011;30:245-52. [Crossref] [PubMed]
- Ferro R, Adamska A, Lattanzio R, et al. GPR55 signalling promotes proliferation of pancreatic cancer cells and tumour growth in mice, and its inhibition increases effects of gemcitabine. Oncogene 2018;37:6368-82. [Crossref] [PubMed]
- Kargl J, Andersen L, Hasenohrl C, et al. GPR55 promotes migration and adhesion of colon cancer cells indicating a role in metastasis. Br J Pharmacol 2016;173:142-54. [Crossref] [PubMed]
- Hasenoehrl C, Feuersinger D, Sturm EM, et al. G protein-coupled receptor GPR55 promotes colorectal cancer and has opposing effects to cannabinoid receptor 1. Int J Cancer 2018;142:121-32. [Crossref] [PubMed]
- Zhou XL, Guo X, Song YP, et al. The LPI/GPR55 axis enhances human breast cancer cell migration via HBXIP and p-MLC signaling. ACTA Pharmacol Sin 2018;39:459-71. [Crossref] [PubMed]
- Andradas C, Blasco-Benito S, Castillo-Lluva S, et al. Activation of the orphan receptor GPR55 by lysophosphatidylinositol promotes metastasis in triple-negative breast cancer. Oncotarget 2016;7:47565-75. [Crossref] [PubMed]
- He D, Wang J, Zhang C, et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer 2015;14:73. [Crossref] [PubMed]


