
Clinicopathological significance and prognostic implication of CD44 and its splice variants (v3 and v6) in colorectal cancer
Introduction
Colorectal cancer (CRC) remains one of the leading causes of mortality in China, as >376,300 newly-diagnosed cases and 191,000 cases of CRC-associated mortality were registered in 2015 (1). The disease is regarded as a classic model for cancer research, since a series of genetic alterations have been reported to be involved in the processes of evolution from normal colonic epithelial cells to invasive cancer (2). At present, it has been demonstrated that most early-stage cases are curable by a single surgery, but the long-term survival for advanced cases remains poor (3). In 2016, a phase II study reported that an immunotherapy agent (pembrolizumab) could result in a marked objective response in a subgroup of patients with deficient mismatch repair; however, this therapy is ineffective for the majority of patients who lack the aforementioned characteristics (4). Notably, a number of studies in recent years indicated a key role of colorectal cancer stem cells (CCSCs) (5-7), as well as their niche, in disease initiation and development; this may suggest the potential of novel therapeutic targets.
CD44 is known as a cell-surface trans-membrane glycoprotein, which includes the standard form CD44s (referred to as CD44) and a series of splice variants (8). In the digestive system, CD44, alone or in combination with other markers, has been identified to be a robust marker for CSCs (defined as “a sub-group of cancer cells in a tumor that possess the capacity for self-renewal and cause the heterogeneous lineages”) (9). However, the prognostic value of CD44 in these types of cancer remains controversial. For example, certain studies have indicated that increased CD44 expression in gastric cancer predicts cancer progression, short recurrence-free survival and poor overall survival (10,11); however, there are also reports of negative and opposite results (12,13). Notably, these discrepancies also exist for CRC (14-16).
It is widely accepted that the role of CD44 variants may not be identical to that of CD44 in cancer (8,17). Although the exact function of these molecules remains largely unknown, a number of CD44 variants have been demonstrated to serve an essential role in the processes of epithelial-mesenchymal transition (EMT) and metastasis in CRC, including CD44 splice variant (v)2, CD44v6 and CD44v8-10 (18-20); however, other variants, including CD44v3, have rarely been studied.
The cancer stem cell niche is considered to contribute to maintaining CSCs by providing a key signal. Previously, c-Kit was reported to play an important role in cancer occurrence (21), also, it can promote chemoresistance and tumor-initiating ability in cancer (22). Targeting c-Kit presented sound treatment effect in many cancers including gastrointestinal stromal tumor (GIST), acute myeloid leukemia and melanoma (21). Interestingly, c-Kit-positive cells are found to be an important element in the intestinal stem cell niche (23) and it has also been demonstrated that over-reactive c-Kit cannot only protect colon cancer cells, but also strengthen the capacity of cell proliferation and invasion (24). Based on these results, it is plausible that c-Kit could be associated with CCSCs. We conducted a retrospective study to explore the expression profile of CD44, CD44v3, CD44v6 and c-Kit in CRC.
Methods
Patient enrollment
Between January 2012 and August 2014, 148 cases were retrospectively collected at Hainan hospital of PLA general hospital. All patients underwent radical surgery via the abdomen or by laparoscopy. Patients who met the following requirements were included: (I) age ≥18 years old; (II) with full clinicopathological data and (III) with regular follow-up records. Patients with pre-operative neoadjuvant therapies, a prior history of CRC, carcinoma in situ or multiple cancers were excluded. Clinicopathological parameters recorded in the present study included sex, age, tumor location (right and left), tumor markers [CEA (cat. no. 35542901) and CA19-9 (cat. no. 34729703); the levels of which in the peripheral blood were tested using a Cobase 601 system (all Roche Diagnostics GmbH) within 1 month prior to surgery], maximum tumor diameter (cut-off, 4 cm) (25), tumor shape, cell differentiation and tumor-node-metastasis (TNM) stage according to the 7th Edition of American Joint Committee on Cancer Staging System. The study protocol was approved by the medical ethics committee of Hainan hospital of PLA general hospital (Approved ID: 301HLFYLL15). Written informed consent was obtained from the patients or their relatives.
Follow-up procedure and definition of progression-free survival (PFS)
Restrictive postoperative follow-up procedures were conducted after surgery at intervals of 3–6 months for the first 2 years and at intervals of 6–12 months for the next 3–5 years (all cases were followed for at least 5 years). PFS was determined from the date of surgery until any sign of progression according to response evaluation criteria in solid tumors: RECIST version 1.1 were noted (26), recurrence by computed tomography or the last date of follow-up.
Immunohistochemical determination of CD44, CD44v3, CD44v6 and c-Kit
Fresh samples from the surgery were immediately fixed in 10% formalin for 30 min at room temperature, washed by phosphate-buffered saline (PBS) (cat. no. 10010023; Thermo Fisher Scientific, Inc.) and embedded in paraffin and processed by standard histological methods and the staining was carried out as previously described (27). In brief, at least 4 consecutive 5-µm slides were cut. After deparaffinization at 60 °C for 1 h in a dry oven, sodium citrate solution (cat. no. E673001; BBI Life Science) was used for antigen retrieval for 20 min then cooled to room temperature. Subsequently 3% hydrogen peroxide was used for blocking for 10 min at room temperature, after washing by PBS, the non-specific antigen blocking was then performed with 5% goat serum for 30 min at room temperature (cat. no. C0265; Beyotime Institute of Biotechnology). The primary antibodies used were: Anti-human-CD44 (dilution 1:100; cat. no. ab157107; Abcam), anti-CD44v3 (dilution 1:50; cat. no. ab34229; Abcam), anti-CD44v6-monoclonal antibody (dilution 1:70; cat. no. FAB3660P; R&D Systems, Inc.) and anti-c-Kit (dilution 1:100; cat. no. ab21539; Abcam). All tissues were incubated with the primary antibodies at 4 °C overnight. By washing with PBS again, the slides were then incubated with the secondary antibody (dilution 1:500, cat. no. ab97033/ab97049; Abcam) for 40 min at room temperature. The negative control was set up via the replacement of primary antibodies with identically diluted 5% non-immunized goat serum (cat. no. C0265; Beyotime Institute of Biotechnology) and a GIST was used as the positive control for c-Kit. Staining was achieved using avidin-biotin-peroxidase complex kits (cat. no. 434423; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. The staining results were evaluated under light microscope at a magnification of ×200 (BX51, Olympus Corporation) by two independent pathologists from Hainan hospital of PLA general hospital, who were blinded to the clinical information, and the results were calculated based on the extent of staining (defined as: 0, no staining; 1, 1–10% staining; 2, 11–50% staining; 3, 51–80% staining; and 4, 81–100% staining) and staining intensity (defined as: 0, blank; 1, weak staining; 2, moderate staining; and 3, strong staining). The expression profiles of all markers were stratified as positive or negative based on a total score ≥3 or <3, respectively, as previously described (28).
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 (SPSS, Inc.). Differences in CD44, CD44v3, CD44v6 and c-Kit levels based on various clinicopathological parameters were estimated by a χ2 test or Fisher’s exact test, if appropriate. Univariate and multivariate analyses were performed using the Cox proportional hazards models. Kaplan-Meier survival curves were used to compare patients with positive and negative expression of the markers, and significant differences were determined using the log-rank test. Two-sided P<0.05 was considered to indicate a statistically significant difference, and all data are presented to two decimal places.
Results
Heterogeneous CD44, CD44v3, CD44v6 and c-Kit expression in CRC
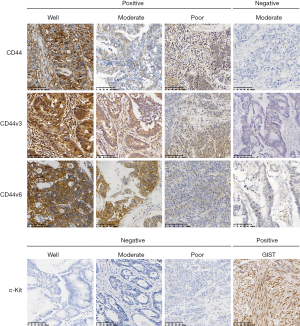
In total, 61 female and 87 male patients were included in the present study. The mean age of the patients was 62.55 years (range, 20–87 years), and the median follow-up time was 30.79 months (range, 3–94 months). A heterogeneous expression profile of the markers was identified in the cohort, and 29 (19.6%), 60 (40.5%) and 66 (44.6%) CRC cases were CD44-, CD44v3- and CD44v6-positive, respectively. No clear positive c-Kit expression was detected in any of the cases. As summarized in Table 1, analysis of clinicopathological features indicated that cell differentiation (P=0.03) and N, M and TNM stages (P=0.04, P<0.01 and P<0.01, respectively) were significantly different between cohorts with or without CD44v3 expression. However, no significant differences were identified among the parameters for CD44, except age (P=0.04), or CD44v6. The immunohistochemical staining patterns (Figure 1) indicated that most of the cancer cells were stained positive for CD44 on the membrane. As for CD44v3, the staining mirrored CD44 on the cell membrane, but some of the cells also exhibited positive expression in the cytoplasm (white arrows). The expression pattern of CD44v6 was similar to that of CD44. In CRC samples, no expression of c-Kit was found in the tumor area, however, it was positive in gastric stromal tumor tissue.
Table 1
| Parameters | No. | CD44 | CD44v3 | CD44v6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos. | Neg. | P | Pos. | Neg. | P | Pos. | Neg. | P | ||||
| Gender | 0.41 | 0.93 | 0.20 | |||||||||
| Female | 61 | 10 | 51 | 25 | 36 | 31 | 30 | |||||
| Male | 87 | 19 | 68 | 35 | 52 | 35 | 52 | |||||
| Age (y) | 0.04* | 0.96 | 0.26 | |||||||||
| ≤60 | 62 | 17 | 45 | 25 | 37 | 31 | 31 | |||||
| >60 | 86 | 12 | 74 | 35 | 51 | 35 | 51 | |||||
| Tumor location | 0.17 | 0.12 | 0.13 | |||||||||
| Left | 73 | 11 | 62 | 25 | 48 | 28 | 45 | |||||
| Right | 75 | 18 | 57 | 35 | 40 | 38 | 37 | |||||
| Maximum tumor diameter (cm) | 0.72 | 0.16 | 0.77 | |||||||||
| ≤4 | 81 | 15 | 66 | 37 | 44 | 37 | 44 | |||||
| >4 | 67 | 14 | 53 | 23 | 44 | 29 | 38 | |||||
| Tumor shape | 0.83 | 0.26 | 0.73 | |||||||||
| Ulcerative | 102 | 21 | 81 | 38 | 64 | 46 | 56 | |||||
| Mass | 42 | 7 | 35 | 19 | 23 | 19 | 23 | |||||
| Infiltrating | 4 | 1 | 3 | 3 | 1 | 1 | 3 | |||||
| Cell differentiation | 1.00 | 0.03* | 0.37 | |||||||||
| Well | 8 | 1 | 7 | 0 | 8 | 2 | 6 | |||||
| Moderate | 122 | 25 | 97 | 53 | 69 | 54 | 68 | |||||
| Poor | 18 | 3 | 15 | 7 | 11 | 10 | 8 | |||||
| T stages | 0.65 | 0.39 | 0.47 | |||||||||
| T1+2 | 19 | 3 | 16 | 6 | 13 | 7 | 12 | |||||
| T3+4 | 129 | 26 | 103 | 54 | 75 | 59 | 70 | |||||
| N stages | 0.53 | 0.04* | 0.19 | |||||||||
| N0 | 74 | 13 | 61 | 24 | 50 | 29 | 45 | |||||
| N1+2 | 74 | 16 | 58 | 36 | 38 | 37 | 37 | |||||
| M stages | 0.79 | 0.00* | 0.36 | |||||||||
| M0 | 115 | 22 | 93 | 40 | 75 | 49 | 66 | |||||
| M1 | 33 | 7 | 26 | 20 | 13 | 17 | 16 | |||||
| TNM stages | 0.47 | 0.00* | 0.18 | |||||||||
| I+II | 65 | 11 | 54 | 18 | 47 | 25 | 40 | |||||
| III+IV | 83 | 18 | 65 | 42 | 41 | 41 | 42 | |||||
*, significant statistical differences. No., number, Pos., positive; Neg., negative.
Association of all parameters, and CD44, CD44v3 and CD44v6 expression with PFS
To further estimate the prognostic value of the markers, the present study examined the association between each marker and PFS (Table 2). The results of univariate Cox analysis indicated that shorter PFS was associated with the following: advanced T stage [hazard ratio (HR), 2.85; 95% CI: 1.38–5.89; P<0.01], N stage (HR 2.59; 95% CI, 1.74–3.86; P<0.01), M stage (HR 2.17; 95% CI, 1.41–3.33; P<0.01), TNM stage (HR 3.06; 95% CI, 2.02–4.65; P<0.01), negative CD44 expression (HR 0.51; 95% CI, 0.35–0.76; P<0.01), positive CD44v3 expression (HR 1.52; 95% CI, 1.04–2.24; P=0.03) and positive CD44v6 expression (HR 1.57; 95% CI, 1.07–2.31; P=0.02). Subsequently, using P=0.05 as a cut-off, multivariate Cox analysis was conducted, which suggested that TNM stage (HR 3.54; 95% CI, 2.31–5.42; P<0.01), negative CD44 expression (HR 0.34; 95% CI, 0.19–0.59; P<0.01), positive CD44v3 expression (HR 1.55; 95% CI, 1.03–2.34; P=0.04) and positive CD44v6 expression (HR 1.59; 95% CI, 1.07–2.38; P=0.02) were independent prognostic factors for CRC (Table 2).
Table 2
| Parameters | Univariable | Multivariable | |||||
|---|---|---|---|---|---|---|---|
| P | HR | 95% CI | P | HR | 95% CI | ||
| Gender | 0.38 | 0.84 | 0.57–1.24 | ||||
| Age | 0.25 | 0.80 | 0.54–1.17 | ||||
| Tumor location | 0.75 | 1.07 | 0.73–1.56 | ||||
| CEA status | 0.47 | 1.15 | 0.79–1.69 | ||||
| CA199 status | 0.13 | 1.41 | 0.91–2.20 | ||||
| Maximum tumor diameter | 0.92 | 1.02 | 0.70–1.50 | ||||
| Tumor shape | 0.26 | 1.24 | 0.86–1.78 | ||||
| Cell differentiation | 0.06 | 1.58 | 0.98–2.53 | ||||
| T stages (T1+2vs. T3+4) | <0.01* | 2.85 | 1.38–5.89 | ||||
| N stages (N0vs. N1+2) | <0.01* | 2.59 | 1.74–3.86 | ||||
| M stages (M0vs. M1) | <0.01* | 2.17 | 1.41–3.33 | ||||
| TNM stages (I+II vs. III+IV) | <0.01* | 3.06 | 2.02–4.65 | <0.01* | 3.54 | 2.31–5.42 | |
| CD44 status (pos. vs. neg.) | <0.01* | 0.51 | 0.35–0.76 | <0.01* | 0.34 | 0.19–0.59 | |
| CD44v3 status (pos. vs. neg.) | 0.03* | 1.52 | 1.04–2.24 | 0.04* | 1.55 | 1.03–2.34 | |
| CD44v6 status (pos. vs. neg.) | 0.02* | 1.57 | 1.07–2.31 | 0.02* | 1.59 | 1.07–2.38 | |
*, significant statistical difference. Pos., positive expression; neg., negative expression.
Prognostic value of CD44, CD44v3 and CD44v6
To further explore the prognostic value of the markers, the present study utilized Kaplan-Meier survival analysis, in which the cumulative survival was defined as the proportion of cases surviving from the date of surgery to the end of the interval. As shown in Figure 2, patients with positive CD44 expression had a significantly longer PFS than patients without (P<0.01). By contrast, patients with positive CD44v3 and CD44v6 expression had a significantly shorter PFS compared with patients without (P=0.03 and P=0.02, respectively; Figure 2).

Discussion
The present study revealed that CD44, CD44v3 and CD44v6 were heterogeneously expressed in CRC, and associations were reported between the positive/negative expression of these markers, particularly CD44v3, and a number of clinicopathological parameters. Patients without CD44 expression, or with CD44v3, v6 expression had a shortened PFS. The results suggest that the prognostic value of CD44 and its splice variants are not unanimous in CRC.
According to the CSC model, CCSCs should be considered to aid cancer development; these cells are resistant to therapy (29). Clusters of these cells in the tumor bulk are an indicator of poor prognosis. CD44 is commonly used as a CSC marker for gastrointestinal types of cancer, particularly colon cancer (14-16). Previous in vitro studies have demonstrated that CD44 can contribute to apoptosis resistance and invasion in colon cancer cells (30,31); however, it is worth noting that the cancer cell lines used in those studies may not recapitulate all features of primary tumors. In fact, a number of clinical reports have indicated that the absence of CD44 (16,32,33), rather than the overexpression of CD44, is associated with adverse events. For example, Hong et al. enrolled 162 patients in a study and found that low CD44 expression is associated with increased tumor recurrence and short DFS (16); similar results have been reported by Qu et al. (32). Additionally, Lugli et al. analyzed 1,420 cases and determined that the loss of CD44 expression is positively associated with advanced T stage, lymph node metastasis and vascular invasion (33); notably, the loss of CD44 is associated with local recurrence (34). In the present study, although no significant differences were observed between the CD44 expression groups and clinicopathological parameters, the role of CD44 expression in predicting shortened PFS was demonstrated, which was consistent with the results of previous studies (16,32).
Previous studies have indicated that alternative splice variants of CD44 are heterogeneously expressed in CRC (13,24). The dynamic balance between CD44 and these variants has essential functions in regulating cancer initiation (35), and in controlling cancer cells undergoing EMT and metastasis (36,37). Interestingly, the majority of studies have indicated an important role of these variants in CRC except CD44v4 (38), for example, high CD44v2 expression is associated with worse prognosis (18), CD44v5 expression is thought related to shorter relapse-free survival (39) but with controversy (40). CD44v8-10 is considered to be associated with lymphatic and venous invasion and liver metastasis (20), and CD44v9 mRNA levels in circulating cancer cells are helpful to predict disease recurrence, prognosis and therapeutic efficacy (41). Notably, CD44v6 may have increased potential as a biomarker than other variants since numerous studies have indicated that CD44v6 overexpression is associated with not only lymph node and distant metastases and treatment response (42-44), but also tumor-associated mortality (45). To the best of our knowledge, there are no published research investigating the expression profile of CD44v3 in CRC; however, in other types of cancer, including gastric (12) and prostate cancer (46), high CD44v3 expression has been revealed to be unimportant for prognostic prediction. In the present study, CD44v6 and CD44v3 expression were found to be independent prognostic factors for patients.
In recent years, the CSC niche has been considered as a therapeutic target for cancer. A previous study indicated that crosstalk between cancer cells and their niche components not only promotes cancer dissemination but also cancer stemness (47). Additionally, c-Kit has been proposed to be a CSC marker; overexpression of c-Kit indicates poor prognosis in a number of cancer types, including ovarian and pancreatic cancer (48,49). Notably, c-Kit-positive cells contribute to the maintenance of stem cells in the normal intestine in animal models (23). A clinical study in CRC demonstrated that c-Kit was over-expressed (50) and that aberrant activation of c-Kit protects colon cancer cells against apoptosis and enhances their invasive potential (24), the underlying mechanisms may involve the regulation of the PI3K/Akt signaling pathway (51). However, positive c-Kit expression in human CRC samples is controversial. Reed et al. conducted a study with 126 samples, and the rate of positive c-Kit expression was only 1.6% (52). The results were further confirmed by another study (53). The underlying reason could be that c-Kit expression is typically restricted to normal interstitial cells, and these cells gradually disappear with malignant transformation of colorectal epithelial cells. In such a scenario, negative immunohistochemical staining can be anticipated in malignant samples (52,53). Nevertheless, the potential association between c-Kit and CCSCs cannot be ruled out since cell signaling can be suppressed, and on depending on the microenvironment at a specific stage of cancer development. The association between c-Kit and CCSCs labeled by markers, such as CD44, CD44v3 and CD44v6, remains to be elucidated in the future.
In conclusion, the present study demonstrated that CD44, CD44v3 and CD44v6 were heterogeneously expressed in CRC, and that these markers could contribute to the prognostic prediction of patients with this disease.
Acknowledgments
Funding: The study was conducted with grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the medical ethics committee of Hainan hospital of PLA general hospital (Approved ID: 301HLFYLL15). Written informed consent was obtained from the patients or their relatives
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer Statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Dienstmann Vermeulen L. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79-92. [Crossref] [PubMed]
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 2009;361:2449-60. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Pd-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Ohata H, Ishiguro T, Aihara Y, et al. Induction of the stem-like cell regulator CD44 by Rho kinase inhibition contributes to the maintenance of colon cancer-initiating cells. Cancer Res 2012;72:5101-10. [Crossref] [PubMed]
- Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 2008;14:6751-60. [Crossref] [PubMed]
- Horst D, Kriegl L, Engel J, et al. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest 2009;27:844-50. [Crossref] [PubMed]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003;4:33-45. [Crossref] [PubMed]
- Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 2006;66:9339-44. [Crossref] [PubMed]
- Senel F, Kokenek Unal TD, Karaman H, et al. Prognostic value of cancer stem cell markers CD44 and ALDH1/2 in gastric cancer cases. Asian Pac J Cancer Prev 2017;18:2527-31. [PubMed]
- Chen Y, Fu Z, Xu S, et al. The prognostic value of CD44 expression in gastric cancer: a meta-analysis. Biomed Pharmacother 2014;68:693-7. [Crossref] [PubMed]
- Setala L, Lipponen P, Tammi R, et al. Expression of CD44 and its variant isoform v3 has no prognostic value in gastric cancer. Histopathology 2001;38:13-20. [Crossref] [PubMed]
- Biddle A, Gammon L, Fazil B, et al. CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal transition. PLoS One 2013;8:e57314. [Crossref] [PubMed]
- Sumiyoshi Y, Yamashita Y, Maekawa T, et al. Expression of CD44, vascular endothelial growth factor, and proliferating cell nuclear antigen in severe venous invasional colorectal cancer and its relationship to liver metastasis. Surg Today 2000;30:323-7. [Crossref] [PubMed]
- Horst D, Kriegl L, Engel J, et al. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest 2009;27:844-50. [Crossref] [PubMed]
- Hong I, Hong SW, Chang YG, et al. Expression of the cancer stem cell markers CD44 and CD133 in colorectal cancer: an immunohistochemical staining analysis. Ann Coloproctol 2015;31:84-91. [Crossref] [PubMed]
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011;11:254-67. [Crossref] [PubMed]
- Ozawa M, Ichikawa Y, Zheng YW, et al. Prognostic significance of CD44 variant 2 upregulation in colorectal cancer. Br J Cancer 2014;111:365-74. [Crossref] [PubMed]
- Masaki T, Goto A, Sugiyama M, et al. Possible contribution of CD44 variant 6 and nuclear beta-catenin expression to the formation of budding tumor cells in patients with T1 colorectal carcinoma. Cancer 2001;92:2539-46. [Crossref] [PubMed]
- Yamaguchi A, Goi T, Taguchi S, et al. Clinical significance of serum levels of CD44 variant exons 8-10 protein in colorectal cancer. J Gastroenterol 1998;33:349-53. [Crossref] [PubMed]
- Abbaspour Babaei M, Kamalidehghan B, Saleem M, et al. Receptor tyrosine kinase (c-Kit) inhibitors: a potential therapeutic target in cancer cells. Drug Des Devel Ther 2016;10:2443-59. [Crossref] [PubMed]
- Chau WK, Ip CK, Mak AS, et al. c-Kit mediates chemoresistance and tumor-initiating capacity of ovarian cancer cells through activation of Wnt/β-catenin-ATP-binding cassette G2 signaling. Oncogene 2013;32:2767-81. [Crossref] [PubMed]
- Rothenberg ME, Nusse Y, Kalisky T, et al. Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology 2012;142:1195-1205.e6. [Crossref] [PubMed]
- Bellone G, Carbone A, Sibona N, et al. Aberrant activation of c-kit protects colon carcinoma cells against apoptosis and enhances their invasive potential. Cancer Res 2001;61:2200-6. [PubMed]
- Santullo F, Biondi A, Cananzi FCM, et al. Tumor size as a prognostic factor in patients with stage IIa colon cancer. Am J Surg 2018;215:71-7. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Yan B, Zhou Y, Feng S, et al. β-Elemene-attenuated tumor angiogenesis by targeting Notch-1 in gastric cancer stem-like cells. Evid Based Complement Alternat Med 2013;2013:268468.
- Ribeiro KB, da Silva Zanetti J, Ribeiro-Silva A, et al. KRAS mutation associated with CD44/CD166 immunoexpression as predictors of worse outcome in metastatic colon cancer. Cancer Biomark 2016;16:513-21. [Crossref] [PubMed]
- Garza-Trevino EN, Said-Fernandez SL, Martinez-Rodriguez HG. Understanding the colon cancer stem cells and perspectives on treatment. Cancer Cell Int 2015;15:2. [Crossref] [PubMed]
- Lakshman M, Subramaniam V, Rubenthiran U, et al. CD44 promotes resistance to apoptosis in human colon cancer cells. Exp Mol Pathol 2004;77:18-25. [Crossref] [PubMed]
- Cho SH, Park YS, Kim HJ, et al. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int J Oncol 2012;41:211-8. [PubMed]
- Qu J, Jiang Y, Liu H, et al. Prognostic value of E-cadherin-, CD44-, and MSH2-associated nomograms in patients with stage II and III colorectal cancer. Transl Oncol 2017;10:121-31. [Crossref] [PubMed]
- Lugli A IG, Hostettler I, Muraro MG, et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer 2010;103:382-90. [Crossref] [PubMed]
- Wong LS, Cantrill JE, Morris AG, et al. Expression of CD44 splice variants in colorectal cancer. Br J Surg 1997;84:363-7. [Crossref] [PubMed]
- Zeilstra J, Joosten SP, van Andel H, et al. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 2014;33:665-70. [Crossref] [PubMed]
- Wielenga VJ, Heider KH, Offerhaus GJ, et al. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res 1993;53:4754-6. [PubMed]
- Biddle A, Gammon L, Fazil B, et al. CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal transition. PLoS One 2013;8:e57314. [Crossref] [PubMed]
- Zalewski B, Stasiak-Barmuta A, Sulkowski S, et al. Glycoprotein CD44 variant 4 expression in tumour epithelial cells of patients with colorectal cancer. Rocz Akad Med Bialymst 2004;49:37-9. [PubMed]
- Vizoso FJ, Fernández JC, Corte MD, et al. Expression and clinical significance of CD44V5 and CD44V6 in resectable colorectal cancer. J Cancer Res Clin Oncol 2004;130:679-86. [Crossref] [PubMed]
- Zalewski B. Levels of v5 and v6 CD44 splice variants in serum of patients with colorectal cancer are not correlated with pT stage, histopathological grade of malignancy and clinical features. World J Gastroenterol 2004;10:583-5. [Crossref] [PubMed]
- Katoh S, Goi T, Naruse T, et al. Cancer stem cell marker in circulating tumor cells: expression of CD44 variant exon 9 is strongly correlated to treatment refractoriness, recurrence and prognosis of human colorectal cancer. Anticancer Res 2015;35:239-44. [PubMed]
- Nihei Z, Ichikawa W, Kojima K, et al. The positive relationship between the expression of CD44 variant 6 and prognosis in colorectal cancer. Surg Today 1996;26:760-1. [Crossref] [PubMed]
- Todaro M, Gaggianesi M, Catalano V, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014;14:342-56. [Crossref] [PubMed]
- Bendardaf R, Lamlum H, Ristamaki R, et al. CD44 variant 6 expression predicts response to treatment in advanced colorectal cancer. Oncol Rep 2004;11:41-5. [PubMed]
- Zhao LH, Lin QL, Wei J, et al. CD44v6 expression in patients with stage II or stage III sporadic colorectal cancer is superior to CD44 expression for predicting progression. Int J Clin Exp Pathol 2015;8:692-701. [PubMed]
- Aaltomaa S, Lipponen P, Ala-Opas M, et al. Expression and prognostic value of CD44 standard and variant v3 and v6 isoforms in prostate cancer. Eur Urol 2001;39:138-44. [Crossref] [PubMed]
- Huang D, Du X. Crosstalk between tumor cells and microenvironment via Wnt pathway in colorectal cancer dissemination. World J Gastroenterol 2008;14:1823-7. [Crossref] [PubMed]
- Yang B, Yan X, Liu L, et al. Overexpression of the cancer stem cell marker CD117 predicts poor prognosis in epithelial ovarian cancer patients: evidence from meta-analysis. Onco Targets Ther 2017;10:2951-61. [Crossref] [PubMed]
- Yasuda A, Sawai H, Takahashi H, et al. The stem cell factor/c-kit receptor pathway enhances proliferation and invasion of pancreatic cancer cells. Mol Cancer 2006;5:46. [Crossref] [PubMed]
- El-Serafi MM, Bahnassy AA, Ali NM, et al. The prognostic value of c-Kit, K-ras codon 12, and p53 codon 72 mutations in Egyptian patients with stage II colorectal cancer. Cancer 2010;116:4954-64. [Crossref] [PubMed]
- Yasuda A, Sawai H, Takahashi H, et al. Stem cell factor/c-kit receptor signaling enhances the proliferation and invasion of colorectal cancer cells through the PI3K/Akt pathway. Dig Dis Sci 2007;52:2292-300. [Crossref] [PubMed]
- Reed J, Ouban A, Schickor FK, et al. Immunohistochemical staining for c-Kit (CD117) is a rare event in human colorectal carcinoma. Clin Colorectal Cancer 2002;2:119-22. [Crossref] [PubMed]
- Sammarco I, Capurso G, Coppola L, et al. Expression of the proto-oncogene c-KIT in normal and tumor tissues from colorectal carcinoma patients. Int J Colorectal Dis 2004;19:545-53. [Crossref] [PubMed]


