
Comprehensive analysis of the expression of chaperonin containing TCP1 subunits (CCTs) and their influence on prognosis in hepatocellular carcinoma
Introduction
Liver cancer, predominantly hepatocellular carcinoma (HCC), mainly arises in the context of chronic liver disease and is the sixth most common cancer and the fourth leading cause of cancer-related death worldwide (1). Even after hepatectomy, the 5-year recurrence rate of HCC patients is as high as 70% (2). Epigenetic modifications and the accumulation of somatic genomic alterations in passenger and driver genes make HCC highly refractory to therapeutic intervention (2). In addition, the lack of diagnostic and prognostic risk biomarkers hinders the identification of high-risk populations that could be used to design clinical trials. The current treatment options for advanced-stage HCC yield only marginal survival benefits and are not cost-effective (3). Thus, the identification of reliable early diagnostic and precise prognostic biomarkers and the development of novel molecular targeted therapeutic strategies for HCC patients are urgently needed.
Chaperonin containing TCP1 (CCT), also known as the t-complex polypeptide 1 ring complex (TRiC), is a group II chaperonin that assists in protein folding in the eukaryotic cytosol (4). It forms a barrel-like structure in which two back-to-back subunit rings surround the central cavity. Each ring consists of eight distinct protein subunits, named CCT1 (TCP1), CCT2, CCT3, CCT4, CCT5, CCT6A, CCT7, and CCT8 (5). CCT was first indicated as a molecular chaperone complex that assists in the folding of the tubulin and cytoskeletal proteins actin (6). It was reported that chaperonin containing TCP1 subunits (CCTs) were markedly upregulated from the G1/S phase transition to the early S phase cell cycle (7). Knockdown of CCTs by siRNA or microinjection of inhibitory monoclonal antibody of CCTs could lead to growth arrest and changes in cell morphology and motility (8). Emerging evidence implicates CCTs in tumorigenesis. Interestingly, several proteins associated with tumorigenesis have been identified as CCT components, and include signal transducer and activator transcription 3 (STAT3), P53, cyclin B and E, and von Hippel-Lindau (VHL) tumor suppressor protein (9).
Previous studies have found that some members of the CCT family are aberrantly expressed and associated with HCC tumorigenesis (10). For instance, CCT3 was upregulated in HCC tissues, and high expression of CCT3 was correlated with poor prognosis in HCC patients. CCT3 depletion suppressed cell proliferation by inducing mitotic arrest, apoptosis, and sensitized HCC cells to vincristine (11). Similarly, upregulated expression of CCT8 was detected in HCC samples compared with adjacent normal liver tissues. The expression of CCT8 was correlated with histologic grade and tumor size, and the high expression of CCT8 was associated with a poor prognosis in patients with HCC. Knockdown of CCT8 inhibited the proliferation and blocked the S-phase entry of HCC cells (12).
Based on these previous studies, CCTs have attracted increasing attention as potential prognostic biomarkers and molecular therapeutic targets for HCC patients. However, the role of distinct CCTs in HCC remains largely unknown. To explore the potential value of distinct CCTs in HCC patients, we conducted a comprehensive analysis to investigate the expression levels, genetic alterations, prognostic value, functional enrichments, and molecular mechanisms of CCTs in HCC by using various large databases.
Methods
ONCOMINE analysis
The ONCOMINE database (www.oncomine.org) contains gene expression data and clinical date of various cancer types. Student’s t-test was used to assess the differential expression levels of CCTs between HCC tissues and normal liver tissues. The cutoff points for statistical significance were a P value of 1E-4 and a fold change of at least 1.5.
The Cancer Genome Atlas (TCGA) database
TCGA (https://www.cancer.gov/) is a landmark cancer genomics program that molecularly characterized over 20,000 primary cancer and matched normal samples spanning 33 cancer types. In the present study, data from 371 HCC patients with CCT mRNA expression information were download. Clinical parameters, including age, gender, AFP, Child-Pugh classification, fibrosis score, pathological grade, and vascular invasion, are summarized in Table S1.
GEPIA dataset analysis
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) is a new tool for analyzing the RNA sequencing expression data from the TCGA and GTEx databases. GEPIA can be used for differential expression analysis, similar gene detection and correlation analysis, and survival analysis (13). The transcript levels of CCTs in 369 HCC tissues and 160 normal liver tissues were visualized through GEPIA. CCTs with high |log2FC| values and low q values (more than 1 or less than 0.01, respectively) were considered differentially expressed genes.
The Kaplan-Meier plotter analysis
The Kaplan-Meier plotter (http://kmplot.com/analysis/) is capable of assessing the effect of 54,000 genes on survival in 21 cancer types. The system includes gene chip and RNA sequencing data sources for the databases including GEO, EGA, and TCGA (14). In this study, the prognostic value of mRNA expression of CCTs in HCC patients was assessed by a Kaplan-Meier survival plot, with the hazard ratios (HRs) with 95% confidence intervals (CIs); a log-rank p-value<0.05 was considered a statistically significant difference.
cBioPortal analysis
The cBioPortal (http://www.cbioportal.org/) is a convenient tool that allows for visualization, analysis, and download large-scale cancer genomics data sets. In our study, the TCGA provisional dataset with mRNA data from 360 HCC patients was selected for analysis. In addition, coexpression and network analyses of CCTs were calculated and visualized.
Functional enrichment and pathway analysis
The Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed using R software (with “GO.db,” “DOSE,” “AnnotationHub,” “cluster profiler,” “ReactomePA” and “org.Hs.eg.db” packages). A P value <0.01 indicated statistical significance. The top 10 pathways were visualized.
Results
Transcriptional levels of CCTs in patients with HCC
Eight CCT subunits have been identified in mammalian cells. We compared the transcriptional levels of CCTs in tumor tissues with those in normal liver tissues by using the ONCOMINE database (Figure 1). The results showed that the mRNA expression of CCT1-CCT8 was upregulated in various types of cancers. We further found that CCT1-CCT8 was upregulated in HCC patients in four datasets. In the Roessler liver 2 dataset (15), CCT1 was upregulated in HCC tissues compared with normal liver tissues (P=6.59E-42; fold change =1.802) (Table 1). CCT2 was overexpressed in HCC samples compared with normal liver tissues in the Roessler liver dataset (15) (P=4.23E-06; fold change =1.831) and the Roessler liver 2 dataset (P=1.33E-53; fold change =2.084) (Table 1). CCT3 was upregulated in HCC tissues in three datasets, including the Roessler liver 2 dataset (P=4.88E-107; fold change =2.978), the Roessler liver dataset (P=7.13E-15; fold change =2.968) and the Wurmbach liver dataset (16) (P=4.60E-08; fold change =2.944) (Table 1). In the Roessler liver 2 dataset, CCT4 was also overexpressed in HCC tissues compared to normal liver tissues (P=5.48E-61; fold change =1.95) (Table 1). CCT5 was found to be more highly expressed in HCC tissues than in normal liver tissues in the Roessler liver 2 dataset (P=2.57E-72; fold change =2.173), the Roessler liver dataset (P=3.72E-09; fold change =2.034) and the Chen liver dataset (17) (P=1.70E-09; fold change =1.548) (Table 1). CCT6A was also upregulated in HCC tissues compared to normal liver tissues in the Roessler liver 2 dataset (P=2.32E-85; fold change =2.897), the Roessler liver dataset (P=2.17E-11; fold change =2.696), and the Wurmbach liver dataset (P=3.08E-05; fold change =2.122) (Table 1). In the Roessler liver 2 dataset, CCT7 was found to be upregulated in HCC tissues compared to normal liver tissues (P=9.00E-41; fold change =1.63) (Table 1). In addition, CCT8 was found to be more highly expressed in HCC tissues than in the normal liver tissues in the Roessler liver dataset (P=1.05E-08; fold change =1.686) and the Roessler liver 2 dataset (P=2.25E-48; fold change =1.619) (Table 1). These results showed that CCTs were upregulated in HCC tissues compared with normal liver tissues.
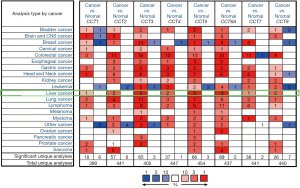
Table 1
| Types of HCC vs. liver | P value | t-test | Fold change | Ref. |
|---|---|---|---|---|
| CCT1 | ||||
| HCC vs. normal | 6.59E-42 | 15.114 | 1.802 | Roessler Liver 2 (15) |
| CCT2 | ||||
| HCC vs. normal | 1.33E-53 | 17.690 | 2.084 | Roessler Liver 2 (15) |
| HCC vs. normal | 4.23E-06 | 5.240 | 1.831 | Roessler Liver (15) |
| CCT3 | ||||
| HCC vs. normal | 4.88E-107 | 31.224 | 2.978 | Roessler Liver 2 (15) |
| HCC vs. normal | 7.13E-15 | 11.906 | 2.968 | Roessler Liver (15) |
| HCC vs. normal | 4.60E-08 | 7.831 | 2.944 | Wurmbach Liver (16) |
| CCT4 | ||||
| HCC vs. normal | 5.48E-61 | 19.313 | 1.950 | Roessler Liver 2 (15) |
| HCC vs. normal | 1.41E-10 | 6.726 | 1.516 | Chen Liver (17) |
| CCT5 | ||||
| HCC vs. normal | 2.57E-72 | 22.528 | 2.173 | Roessler Liver 2 (15) |
| HCC vs. normal | 3.72E-09 | 7.353 | 2.034 | Roessler Liver (15) |
| HCC vs. normal | 1.70E-09 | 6.228 | 1.548 | Chen Liver (17) |
| CCT6A | ||||
| HCC vs. normal | 2.32E-85 | 24.642 | 2.897 | Roessler Liver 2 (15) |
| HCC vs. normal | 2.17E-11 | 8.994 | 2.696 | Roessler Liver (15) |
| HCC vs. normal | 3.08E-05 | 5.203 | 2.122 | Wurmbach Liver (16) |
| CCT7 | ||||
| HCC vs. normal | 9.00E-41 | 14.791 | 1.630 | Roessler Liver 2 (15) |
| CCT8 | ||||
| HCC vs. normal | 1.05E-08 | 7.071 | 1.686 | Roessler Liver (15) |
| HCC vs. normal | 2.25E-48 | 16.512 | 1.619 | Roessler Liver 2 (15) |
Note: P<0.05 is considered statistical significant. HCC, hepatocellular carcinoma; CCT, chaperonin containing TCP1 subunit.
Relationship between the mRNA expression levels of CCTs and clinicopathological parameters in patients with HCC
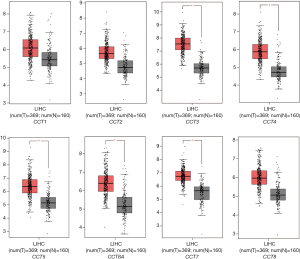
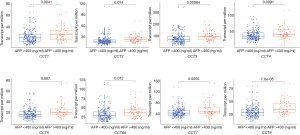
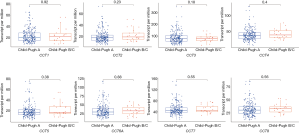
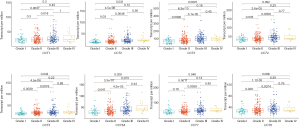
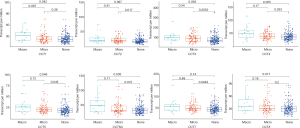
We compared the mRNA expression levels of CCTs between HCC and normal liver tissues by using the GEPIA online analysis tool. The results indicated that the mRNA expression levels of CCT3, CCT4, CCT5, CCT6A, and CCT7 were significantly upregulated, but no significant differences were found in the mRNA expression of CCT1, CCT2, and CCT8 in HCC tissues compared with normal liver tissues (Figure 2). We further analyzed the relationship between the mRNA expression levels of CCTs and alpha-fetoprotein (AFP), Child-Pugh classification, pathological grade, and vascular invasion of patients with HCC (Figures 3-6). The results showed that a higher AFP serum level (the cutoff point was 400 ng/mL) tended to correlate with a higher expression of CCT1-CCT8 (Figure 3). No relationship was found between the Child-Pugh classification and the mRNA expression levels of CCT1-CCT8 (Figure 4). In terms of the pathological grade, the results showed that a higher CCT1-CCT8 expression tended to correlate with a poorer pathological grade when comparing the expression levels between grade I or grade II and grade III. No relationship was found between grades II and IV, possibly due to the small sample size of the grade IV data (Figure 5). In addition, CCT2, CCT3, CCT4, CCT5, CCT6, and CCT7 were upregulated in HCC patients with microvascular invasion compared to patients with nonvascular invasion. Only CCT1, CCT5, and CCT6A were found to be upregulated in HCC patients with macrovascular invasion compared to patients with nonvascular invasion, which might also be due to the small sample of HCC patients with macrovascular invasion. No relationships were found involving CCT expression levels between macro- and microvascular invasion in patients with HCC (Figure 6). These results indicated that the mRNA expression levels of CCTs correlated with the prognosis of HCC.
The prognostic value of CCTs in patients with HCC
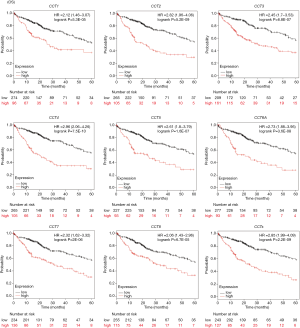
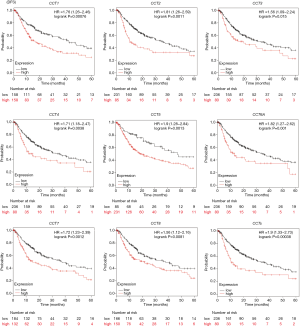
Kaplan-Meier analysis was performed to determine the potential prognostic value of CCTs in HCC patients. As shown in Figure 7, higher mRNA expression of CCT1 (HR =2.12, 95% CI: 1.46–3.07, and P=5.3E-05), CCT2 (HR =2.82, 95% CI: 1.96–4.06, and P=5.2E-09), CCT3 (HR =2.45, 95% CI: 1.7–3.53, and P=6.8E-07), CCT4 (HR =2.96, 95% CI: 2.06–4.26, and P=7.5E-10), CCT5 (HR =2.61, 95% CI: 1.8–3.79, and P=1.6E-07), CCT6A (HR =2.73, 95% CI: 1.88–3.95, and P=3.6E-08), CCT7 (HR =2.32, 95% CI: 1.62–3.32, and P=2E-06), and CCT8 (HR =2.06, 95% CI: 1.43–2.96, and P=6.7E-05) were significantly associated with shorter overall survival (OS) of HCC patients. We further explored the relationship between the combined mRNA expression of all eight CCT subunits (CCTs) and the prognosis of HCC patients. The results showed that the higher combined mRNA expression of the eight CCT subunits (HR =2.85, 95% CI: 1.99–4.09, and P=2.2E-09) was also associated with a shorter OS in patients with HCC. In addition, we further determined the relationship between the mRNA expression of distinct CCTs and the disease-free survival (DFS) of patients with HCC. As shown in Figure 8, a higher mRNA expression of CCT1 (HR =1.76, 95% CI: 1.26–2.46, and P=0.00076), CCT2 (HR =1.81, 95% CI: 1.26–2.59, and P=0.0011), CCT3 (HR =1.56, 95% CI: 1.09–2.24, and P=0.015), CCT4 (HR =1.71, 95% CI: 1.18–2.47, and P=0.0038), CCT5 (HR =1.9, 95% CI: 1.28–2.84, and P=0.0013), CCT6A (HR =1.82, 95% CI: 1.27–2.62, and P=0.001), CCT7 (HR =1.72, 95% CI: 1.23–2.39, and P=0.0012), CCT8 (HR =1.56, 95% CI: 1.12–2.16, and P=0.0081) and combined CCTs (HR =1.9, 95% CI: 1.33–2.73, and P=0.00038) were significantly associated with DFS of HCC patients. These results indicated that the mRNA expression of all eight CCTs (CCT1-CCT8) was significantly correlated with the HCC patient prognosis. They may serve as potential biomarkers for the prognosis of HCC patients.
Genetic alteration of CCTs and their correlation with the prognosis of HCC patients
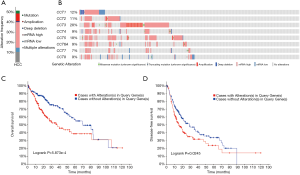
To further explore the prognostic value of CCTs, we analyzed genetic alterations in CCTs and their association with OS and DFS in HCC patients. The results showed that the genes were altered in 51.39% of 360 HCC patients (Figure 9A), including 2.78% (10 patients), amplification 7.78% (28 patients), deep deletion 0.83% (3 patients), mRNA high 25.56% (92 patients), mRNA low 4.17% (15 patients) and multiple alterations 10.28% (37 patients) (Figure 9B). CCT3 (28%), CCT5 (18%), and CCT1 (12%) were ranked as the three genes with the most genetic alterations. Furthermore, we investigated the prognostic value of query CCTs in HCC patients with or without alterations by Kaplan-Meier analysis. The results showed that a genetic alteration in the query CCTs was associated with shorter OS (P=5.873E-4, Figure 9C) and DFS (P=0.0245, Figure 9D) of HCC patients. These results indicated that CCT alteration frequency was associated with poorer prognosis in patients with HCC.
Coexpression and interaction analyses of CCTs and their frequently altered neighboring genes in HCC patients
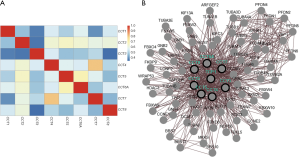
We analyzed the correlations of CCTs with each other by using the cBioPortal online tool. The results showed that CCT1 was significantly and negatively correlated with CCT3 and CCT7. CCT2 was significantly and positively correlated with CCT4, CCT5, CCT6, CCT7, and CCT8 but negatively correlated with CCT3. CCT3 was significantly and negatively correlated with CCT1, CCT2, and CCT8. CCT4 was significantly and positively correlated with CCT2 and CCT8. CCT5 was significantly and positively correlated with CCT2 and CCT6A. CCT6A was significantly and positively correlated with CCT2 and CCT5. CCT7 significantly and positively correlated with CCT4 but negatively correlated with CCT1. CCT8 was significantly and positively correlated with CCT2 but negatively correlated with CCT3 (Figure 10A). Moreover, we further constructed an interaction network for CCTs and their 69 most related neighboring genes (Figure 10B). The results showed that the PI3K/AKT signaling pathway-related genes, including CCNE2, CCNE1, GNB2, GNB1, TP53, GNB5, GNB4, and GNB3, were significantly related to CCT alterations, suggesting that CCTs might serve as therapeutic targets for patients with HCC.
Functional enrichment and signaling pathways affected by CCTs in patients with HCC
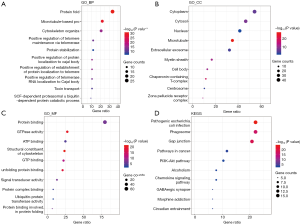
GO analysis predicts three functions, including biological processes (BPs), cellular components (CPs), and molecular functions (MFs) of CCTs and their 69 neighboring genes. The top 10 of those three main functions are shown in Figure 11A,B,C. CCTs and their neighboring genes were enriched in CPs of the cytoplasm, cytosol, nucleus, microtubule, and extracellular exosomes, assisting in protein folding, microtubule-based processes, cytoskeleton organization, positive regulation of telomere maintenance via telomerase, and protein stabilization. The molecular functions for these genes were mainly protein binding, GTPase activity, ATP binding, structural constituent of the cytoskeleton, GTP binding, and signal transducer activity. Moreover, the top 10 KEGG pathways for CCTs and their neighboring 69 genes are shown in Figure 11D. Among these pathways, the pathways in cancer, PI3K/AKT signaling pathway, chemokine signaling pathway, gap junction, phagosome, and alcoholism terms were involved in the development, progression, and metastasis of HCC.
Discussion
CCTs are key molecular complexes that assist in the folding of proteins to produce energy stability and functionally competent protein conformations, which play an essential role in cell growth and survival. Many of the key regulators of cell growth and differentiation have been identified as clients of CCTs, suggesting that CCTs are potential mediators of oncogenesis (18). P53 regulates cell cycle arrest and apoptosis and hence functions as a tumor suppressor. A previous report showed that the folding of wild-type P53 is promoted by the interaction with CCTs. Genetic alterations of CCTs result in the accumulation of misfolded P53, leading to a reduction in P53-dependent gene expression and acquiring an ability to promote invasion and metastasis (19). The functions of CCTs in the tumorigenesis and prognosis of various tumors have been partially explored. However, the patterns of expression and precise roles of different CCTs in HCC are largely unknown. In this study, we attempted to systematically investigate the expression patterns, genetic alterations, prognostic values, functional enrichment, and molecular mechanisms of CCTs in HCC.
CCT1 was upregulated in breast cancer tissues and correlated with the survival of patients with breast cancer. An in vitro study further identified that CCT1 was regulated by driver oncogene activation of phosphatidylinositide 3-kinase (PI3K) signaling and necessary for the growth and survival of breast cancer cells (20). In our study, the ONCOMINE and GEPIA datasets revealed that the expression of CCT1 was upregulated in HCC tissues compared with normal liver tissues. Moreover, CCT1 mRNA expression was correlated with AFP, pathological grade, and macro- and microvascular invasion but was not related to the Child-Pugh classification. Kaplan-Meier analysis indicated that higher mRNA expression of CCT1 was significantly associated with shorter OS and DFS of patients with HCC. These results suggest that CCT1 acts as an oncogene.
CCT2 functions as part of the multimeric CCT complex to produce proteins required for cell growth and division. CCT2 was shown to have upregulated expression in invasive ductal carcinomas compared to cancer adjacent tissues, and this trend increased with breast cancer stage. Higher expression or genetic alternation of CCT2 was correlated with a poorer prognosis in breast cancer (21). Moreover, similar results showed that CCT2 was overexpressed in colorectal cancer (22) and gallbladder carcinoma (23), and high levels of CCT2 were related to a poorer clinical outcome in those patients. Another study was performed to indicate that the disruption of the CCT2/β-tubulin complex could selectively kill the CCT2-overexpressing cancer cells through MAPK activation (9). In this study, CCT2 was found to be more highly expressed in HCC tissues than in normal liver tissues. In addition, CCT2 mRNA expression was correlated with AFP, pathological grade, and macro- and microvascular invasion but was not related to the Child-Pugh classification. Kaplan-Meier analysis showed that high CCT2 expression was significantly associated with the shorter OS and DFS of patients with HCC. These results suggest that CCT2 is a potential target for therapeutic intervention and serves as a prognostic biomarker.
CCT3 is one of the subunits of the CCT complex, which plays a critical role in maintaining cellular proteostasis. A previous study indicated that the expression levels of CCT3 mRNA and protein were upregulated in HCC cells and that high levels of CCT3 were correlated with a poor prognosis in HCC patients. Knockdown of CCT3 suppressed proliferation, invasion, and induced apoptosis in HCC cell lines (24). Another report demonstrated that CCT3 is was required for cell proliferation and spindle integrity during mitosis. CCT3 depletion sensitized HCC cells to the microtubule destabilizing drug vincristine (11). In agreement with previous studies, our observations indicated that CCT3 was overexpressed in HCC tissues compared with normal liver tissues. Moreover, CCT3 expression was associated with AFP, pathological grade, and macro- and microvascular invasion. Survival analysis showed that higher CCT3 expression was significantly associated with a poorer prognosis in patients with HCC. Combined with these previous results, CCT3 may serve as a prognostic biomarker and potential therapeutic target.
Wound healing assays indicated that the CCT4 monomer expression level influences directional cell migration and is associated with the assembled cytoskeleton (25). In our study, the expression of CCT4 was upregulated in HCC samples compared to normal liver tissues and correlated with AFP, pathological grade, and macro- and microvascular invasion. Survival analysis showed that increased CCT4 expression was significantly correlated with shorter OS and DFS in patients with HCC. These results suggest that CCT4 is a potential prognostic biomarker.
CCT5 was significantly overexpressed in primary colorectal cancer and correlated with Dukes’ stage and colorectal cancer progression (22). Furthermore, CCT5 was significantly upregulated in p53-mutated breast tumor samples compared to p53-wild-type breast tumor samples. Upregulated expression of CCT5 was associated with a low response rate to docetaxel, and CCT5 knockdown resulted in a significant enhancement of docetaxel-induced apoptosis in MCF-7 breast cancer cells (26). In this study, the expression of CCT5 was upregulated in HCC tissues compared with normal liver tissues and correlated with AFP, pathological grade, and macro- and microvascular invasion. Survival analysis showed that increased CCT5 expression was significantly correlated with a poorer prognosis in patients with HCC. These results suggest that CCT5 may act as a prognostic biomarker and potential therapeutic target.
CCT6A encodes a molecular chaperone of the CCT complex that mediates protein folding in the eukaryotic cytosol. Differential expression analysis showed that CCT6A was significantly upregulated in almost all lung tumor tissues. Analyses based on genes and pathways revealed that CCT6A might contribute to the survival of patients with non-small-cell lung carcinoma (NSCLC) (27). Another study identified that CCT6A was a specific blocker of SMAD2, and in vivo studies showed that silencing CCT6A resulted in the efficient suppression of TGF-β-mediated metastasis in NSCLC cells and prolonged the survival of tumor-bearing mice (28). In this study, the expression of CCT6A was upregulated in HCC tissues compared to normal liver tissues and was correlated with AFP, pathological grade, and macro- and microvascular invasion but was not related to Child-Pugh classification. Survival analysis indicated that increased expression of CCT6A was significantly correlated with shorter OS and DFS of patients with HCC. These results suggest a new potential strategy for specifically targeting tumor-promoting signaling in cancer.
siRNA-mediated depletion of CCT7 inhibited the accumulation of α-SMA protein in Dupuytren’s contracture (DC) cells and significantly decreased the contractility of treated DC cells (29). In this study, the expression of CCT7 was upregulated in HCC tissues compared with the expression in normal liver tissues and correlated with AFP, pathological grade, and macro- and microvascular invasion but not with the Child-Pugh classification. Survival analysis found that increased CCT7 expression was significantly correlated with shorter OS and DFS of HCC patients. These findings suggest that CCT7 may serve as a potential prognostic biomarker of HCC patients.
CCT8 plays an essential role in the proliferation, migration, and invasion of several types of cancer. In colorectal cancer, glioma (30), esophageal squamous cell carcinoma (ESCC) (31), and HCC (12), the expression of CCT8 is upregulated in tumor tissues compared to normal tissues, and the high expression of CCT8 is significantly associated with poor prognosis. Further studies showed that CCT8-siRNA significantly decreased the proliferation and invasion capacity of glioma cells (30). In addition, CCT8 promoted ESCC cell migration and invasion by regulating alpha-actin and beta-tubulin expression, and overexpression of CCT8 is correlated with poor prognosis and cisplatin resistance in ESCC patients (31). Interestingly, knockdown of CCT8 blocked S-phase entry and suppressed cell proliferation in HCC cells (12). Consistent with previous results, our study showed that CCT8 was upregulated in HCC compared to that in normal liver tissues and correlated with AFP, pathological grade, and macro- and microvascular invasion but was not related to the Child-Pugh classification. The survival analysis showed that increased CCT8 expression was significantly correlated with shorter OS and DFS of patients with HCC. These findings suggest that CCT8 might be an oncogene and potential therapeutic strategy for the treatment of HCC.
To further explore the prognostic value of CCTs, we analyzed the genetic alterations in CCTs and their correlations with OS and DFS in HCC patients. The results showed that genetic alterations occurred in 51.39% of HCC patients. Interestingly, patients with genetic alterations in query CCTs had a poorer prognosis than those without genetic alterations. We constructed a functional enrichment and interaction network for eight CCTs and their 69 neighboring genes. The results indicated that the molecular functions of these genes were protein binding, GTPase activity, ATP binding, structural constituents of the cytoskeleton, GTP binding, and signal transducer activity. Furthermore, the pathways in cancer, PI3K/AKT signaling pathway, chemokine signaling pathway, gap junction, phagosome, and alcoholism terms were concerned with tumorigenesis, development, and metastasis of HCC, which offers clues into the rational development of multitargeted and CCT-mediated targeted therapies for HCC patients.
Conclusions
In summary, our results indicated that the mRNA expression levels of CCTs were significantly upregulated and obviously and positively associated with the AFP, pathologic grade, and macro- and microvascular invasion in HCC patients. Furthermore, aberrant expression and genetic alterations of CCTs were correlated with the prognosis of HCC patients. Our findings suggest that CCTs may serve as potential prognostic biomarkers and therapeutic targets in HCC patients. These results may be beneficial for a better understanding of the molecular mechanism of HCC and may be useful to develop tools for accurate HCC prognosis and for promoting the development of CCT-mediated drugs for HCC treatment. However, the development of new methodologies and further research are warranted to increase our understanding of the role of CCTs in HCC development and progression, which could be exploited to develop therapeutic strategies for patients with HCC.
Table S1
| Clinical characteristic | Number of patients | % |
|---|---|---|
| Age (years) | ||
| <60 | 169 | 45.55 |
| ≥60 | 201 | 54.18 |
| Gender | ||
| Female | 121 | 32.61 |
| Male | 250 | 67.39 |
| Child-Pugh | ||
| A | 217 | 58.49 |
| B/C | 22 | 5.93 |
| Fibrosis score | ||
| 0–4 | 133 | 35.85 |
| 5–6 | 79 | 21.29 |
| Grade | ||
| Well differentiated | 55 | 14.82 |
| Moderately differentiated | 177 | 47.71 |
| Poorly differentiated | 122 | 32.88 |
| Undifferentiated | 12 | 3.23 |
| Vascular cell type | ||
| None | 206 | 55.53 |
| Macro | 16 | 4.31 |
| Micro | 93 | 25.07 |
| AFP (ng/mL) | ||
| <400 | 213 | 53.41 |
| >400 | 65 | 17.52 |
| Total | 371 | 100 |
HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.20). The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Parikh ND, Singal AG, Hutton DW. Cost effectiveness of regorafenib as second-line therapy for patients with advanced hepatocellular carcinoma. Cancer 2017;123:3725-31. [Crossref] [PubMed]
- Kubota S, Kubota H, Nagata K. Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc Natl Acad Sci U S A 2006;103:8360-5. [Crossref] [PubMed]
- Sternlicht H, Farr GW, Sternlicht ML, et al. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci U S A 1993;90:9422-6. [Crossref] [PubMed]
- Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biochem 1995;230:3-16. [Crossref] [PubMed]
- Yokota S, Yanagi H, Yura T, et al. Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem 1999;274:37070-8. [Crossref] [PubMed]
- Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res 2006;312:2309-24. [Crossref] [PubMed]
- Liu YJ, Kumar V, Lin YF, et al. Disrupting CCT-beta: beta-tubulin selectively kills CCT-beta overexpressed cancer cells through MAPKs activation. Cell Death Dis 2017;8:e3052. [Crossref] [PubMed]
- Yokota S, Yamamoto Y, Shimizu K, et al. Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperones 2001;6:345-50. [Crossref] [PubMed]
- Zhang Y, Wang Y, Wei Y, et al. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett 2016;372:101-9. [Crossref] [PubMed]
- Huang X, Wang X, Cheng C, et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS 2014;122:1070-9. [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202-12. [Crossref] [PubMed]
- Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938-47. [Crossref] [PubMed]
- Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929-39. [Crossref] [PubMed]
- Roh SH, Kasembeli M, Bakthavatsalam D, et al. Contribution of the Type II Chaperonin, TRiC/CCT, to Oncogenesis. Int J Mol Sci 2015;16:26706-20. [Crossref] [PubMed]
- Trinidad AG, Muller PA, Cuellar J, et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell 2013;50:805-17. [Crossref] [PubMed]
- Guest ST, Kratche ZR, Bollig-Fischer A, et al. Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp Cell Res 2015;332:223-35. [Crossref] [PubMed]
- Bassiouni R, Nemec KN, Iketani A, et al. Chaperonin Containing TCP-1 Protein Level in Breast Cancer Cells Predicts Therapeutic Application of a Cytotoxic Peptide. Clin Cancer Res 2016;22:4366-79. [Crossref] [PubMed]
- Coghlin C, Carpenter B, Dundas SR, et al. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol 2006;210:351-7. [Crossref] [PubMed]
- Zou Q, Yang ZL, Yuan Y, et al. Clinicopathological features and CCT2 and PDIA2 expression in gallbladder squamous/adenosquamous carcinoma and gallbladder adenocarcinoma. World J Surg Oncol 2013;11:143. [Crossref] [PubMed]
- Cui X, Hu ZP, Li Z, et al. Overexpression of chaperonin containing TCP1, subunit 3 predicts poor prognosis in hepatocellular carcinoma. World J Gastroenterol 2015;21:8588-604. [Crossref] [PubMed]
- Echbarthi M, Vallin J, Grantham J. Interactions between monomeric CCTdelta and p150(Glued): A novel function for CCTdelta at the cell periphery distinct from the protein folding activity of the molecular chaperone CCT. Exp Cell Res 2018;370:137-49. [Crossref] [PubMed]
- Ooe A, Kato K, Noguchi S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res Treat 2007;101:305-15. [Crossref] [PubMed]
- Zhu M, Geng L, Shen W, et al. Exome-Wide Association Study Identifies Low-Frequency Coding Variants in 2p23.2 and 7p11.2 Associated with Survival of Non-Small Cell Lung Cancer Patients. J Thorac Oncol 2017;12:644-56. [Crossref] [PubMed]
- Ying Z, Tian H, Li Y, et al. CCT6A suppresses SMAD2 and promotes prometastatic TGF-beta signaling. J Clin Invest 2017;127:1725-40. [Crossref] [PubMed]
- Satish L, O'Gorman DB, Johnson S, et al. Increased CCT-eta expression is a marker of latent and active disease and a modulator of fibroblast contractility in Dupuytren's contracture. Cell Stress Chaperones 2013;18:397-404. [Crossref] [PubMed]
- Qiu X, He X, Huang Q, et al. Overexpression of CCT8 and its significance for tumor cell proliferation, migration and invasion in glioma. Pathol Res Pract 2015;211:717-25. [Crossref] [PubMed]
- Yang X, Ren H, Shao Y, et al. Chaperonin-containing T-complex protein 1 subunit 8 promotes cell migration and invasion in human esophageal squamous cell carcinoma by regulating alpha-actin and beta-tubulin expression. Int J Oncol 2018;52:2021-30. [PubMed]


