
Circulating guanylyl cyclase C (GCC) mRNA is a reliable metastatic predictor and prognostic index of colorectal cancer
Introduction
Colorectal cancer (CRC) is the third-leading cause of cancer-related death worldwide due to its tendency to metastasize to distant organs (1). In spite of effective treatments like standard radical surgery and adjuvant therapies, circulating tumor cells (CTCs) still lead to unexpected clinical metastases during the follow-up process, and do so partially in early-stage CRC patients (2,3). CTC has been shown to play a crucial role in tumor metastasis from the local primary site to distant target organs (4,5). It is also a valuable carrier for both diagnosing micro-metastasis and predicting the patients’ prognosis in the advanced stage of the tumor (6,7). CTCs are currently considered to be reliable predictors of the overall survival (OS) and progression-free survival (PFS) in local advanced CRC patients with metastasis (8). For instance, Rahbari demonstrated that the detection of CTCs indicated poor prognosis in CRC patients (9). However, the value of CTC detection in early-stage CRC patients is still unclear due to the low collection efficiency of CTC, and the fact it is a non-specific tumor biomarker with no standardized cutoff value (10,11). Among all the CTC enrichment and isolation techniques, the CellSearch System Platform is superior in assessing the circulating tumor burden in metastatic CRC patients, while quantitative real-time PCR (qRT-PCR) is highly sensitive, therefore commonly used in early-stage CRC patients (12). In non-metastatic CRC patients, qRT-PCR might be optimal for the detection of CTCs; however, there remains both a specific marker and a standard cutoff value for predicting metastasis and survival are lacking (5,13).
Guanylyl cyclase C (GCC) is the transmembrane receptor of the diarrheagenic bacterial heat-stable enterotoxin that is selectively expressed in intestinal epithelial cells from the duodenum to the rectum (14,15). As a member of the guanylyl cyclase family, GCC expression persists only in normal intestinal cells and intestinal tumor cells and their metastases but seldom does so in extra-intestinal tissues and tumors, indicating its potential application as a tumor biomarker for the identification of CRC (14,16,17). GCC is also an intestine-restricted protein and thus suitable to act as a cancer mucosa antigen (CMA) of CRC in clinical practice (14,18). Furthermore, like carcinoembryonic antigen (CEA) and cytokeratin 20 (CK20), circulating GCC mRNA can be detected as a CTC-associated biomarker in metastatic CRC patients (19,20). A comparison of multiple epithelial cell biomarkers has demonstrated that GCC is one of the most sensitive and specific biomarkers for the detection of CRC (19,21). Although CTC cell count >3 has been established as the cutoff value for clinical use in the metastatic setting of CRC patients, the value of mRNA as the cutoff point has not yet been clarified in early-stage CRC patients (22-24).
Therefore, this study aimed to investigate circulating GCC mRNA and its association with clinicopathological characteristics, distant organ metastasis, and long-term survival in stage I–III CRC patients.
Methods
A total of 160 CRC patients at stage I–III, who underwent surgery at the Surgical Department of Colorectal Cancer in Zhejiang Cancer Hospital (Hangzhou, China), were recruited from December 2012 to December 2014, and retrospectively analyzed in December 2017. Patients with stage IV secondary tumors outside of the colorectal tract or with benign intestinal tumors were excluded from the study. Peripheral venous blood was preoperatively obtained during clinical staging for the detection of the mRNA level. The first 2 mL of blood was discarded in order to minimize the possibility of false-positives, and the subsequent 5 mL of blood was collected into EDTA-containing vacutainer tubes. Pathological examinations were performed when all the colorectal tumors were surgically removed. Blood samples from five healthy donors and five cases of benign intestine tumors were taken as negative controls. After collection, all samples were processed within 2 hours, immediately stored in cryovials, frozen in liquid nitrogen, and preserved at −80 °C until further processing. Total RNA was extracted from peripheral blood using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. GCC mRNA was prepared from 2 µg of total RNA using the corresponding quantitative PCR kits (Shanghai Jiusheng Medical Co. Ltd., China), according to the manufacturer’s instructions. Primers were designed as described previously (25). The cutoff value of 500 copies/µL for GCC mRNA was based on the manufacturer’s instructions. Consequently, based on the deduced distribution curve, the highest Youden index was observed at 375 copies/µL for GCC mRNA by disease-free survival (DFS) analysis, while 322 copies/µL were observed for GCC mRNA by OS analysis (refer to GCC supplementary ROC curve). The total mRNA levels were categorized into 25 groups according to the cutoff value at 0, 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,500, 2,000, 2,500, 3,000, 3,500, 4,000, 4,500, 5,000, 6,000, 7,000, 8,000, 9,000, and 10,000 copies/µL in order to investigate the correlation of different mRNA cutoff values with the DFS and OS of CRC patients. The two subgroups with higher or lower cutoff values than those of GCC mRNA were summarized and analyzed statistically.
All patients were followed up at periodic intervals (every 3 months for 2 years, every 6 months between 2 and 5 years, and annually after 5 years post-operation). In order to ensure the patients were alive and to learn about the development of any distant organ metastasis, the follow-up activities were performed by means of computed tomography (CT) scan, tumor biomarker detection, colonoscopy, in addition to the letter, telephone, and comprehensive review. All the study protocols were approved by the Institutional Review Board, and informed consent was obtained from all the participants. The median follow-up time was 51.98 (range from 6 to 60) months, and the follow-up ended on December 30, 2017. OS and DFS were defined as described previously. The distribution of GCC mRNA levels higher than the cutoff value was summarized as n (%) according to a given subject’s demographics and clinical characteristics. The series of cutoff mRNA copy numbers were stratified by clinical staging and assessed with DFS and OS. Univariate Cox-regression model analyses were used to evaluate the correlation of DFS or OS with the patients’ demographics and clinical characteristics. These clinicopathological characteristics showed a significant correlation with DFS and OS in the univariate analysis, which led to a multivariate Cox-regression model analysis equivalent to Backward Stepwise [conditional likelihood ratio (LR)] being conducted. Furthermore, the hazard ratio (HR) for survival was estimated by multivariate analysis according to the mRNA copy numbers adjusted by other characteristics. The OS or DFS for GCC mRNA levels and clinicopathological variables were evaluated via Kaplan-Meier curves with the log-rank test. The GCC mRNA of CRC patients with and without distal metastasis during follow-up was compared via t-test. A multivariate logistic regression-based nomogram was made for predicting survival in CRC patients. All statistical tests were two-sided with a 95% confidence interval (CI); P<0.05 was considered as statistically significant. All statistical analyses were performed with PASW statistics software version 23.0 (SPSS Inc., Chicago, IL, USA).
Results
Baseline characteristics and GCC mRNA levels of CRC patients and healthy donors
The study population comprised 101 (63.1%) males and 59 (36.9%) females, with 60 (37.5%) colon carcinomas and 100 (62.5%) rectal carcinomas. The mean age of the CRC patients was 66.7 years old. According to the Union for International Cancer Control (UICC) Classification of CRC, 31 patients (19.4%) were at stage I, 58 (36.3%) at stage II, and 71 (44.4%) at stage III. A total of ten blood samples from healthy donors and benign intestinal tumors were collected, comprising 6 males and 4 females, with an average age of 60.4 years. GCC mRNA was negative in the ten blood samples (0/10), which was significantly lower than that in the study population of 26.9% (43/160).
Univariate regression analysis of clinicopathological factors for DFS and OS
As shown in Table S1, DFS decrease was significantly associated with the presence of tumor emboli in vessels, lymph node metastases, mesenteric root lymph node metastases, ulcerative pathological type, low differentiation type, stage III, and high CA199 values (all P<0.05). Meanwhile, tumor size (>5 cm), low differentiation type, tumor emboli in vessels, lymph node metastases, and mesenteric root lymph node metastases were found to be significantly associated with reduced OS (all P<0.05).
Correlation of GCC mRNA level with distal metastasis
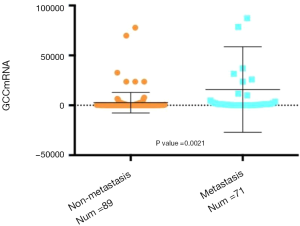
The comparison of circulating GCC mRNA level was carried out between the CRC patients who ultimately developed distal metastasis during their follow-up and those without distal metastasis. As shown in Figure 1, the copy number of GCC mRNA in the metastasis subgroup was significantly higher than that in the non-metastasis subgroup (P=0.0021).
Univariate regression analysis of GCC mRNA for DFS and OS
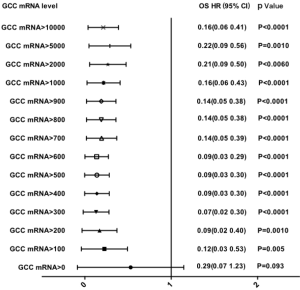
The 25 groups from the subgroup analysis with a cutoff value between 0 and 10,000 copies/µL demonstrated that the level of GCC mRNA with a cutoff value >50 copies/µL was significantly associated with reduced OS while the levels with a cutoff value >100 copies/µL were significantly associated with DFS decrease (see Table S2). Furthermore, the correlation of OS and DFS with the GCC mRNA level was analyzed based on stage stratification (see Table S3). GCC mRNA over baseline (>100 copies/µL) showed a significant correlation with OS and DFS in the stage II subgroup. Meanwhile, a significant correlation of reduced OS with GCC mRNA with a cutoff value >1,500 copies/µL was observed in the stage I subgroups. In the stage III subgroups, GCC mRNA levels from 200 to 900 copies/µL and from 50 to 4,500 copies/µL were significantly associated with a reduction of both DFS and OS.
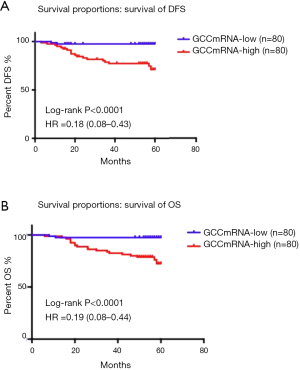
Survival proportion analysis of GCC mRNA subgroups (lower than the median value of copy numbers) showed better DFS and OS than those of GCC mRNA in the high subgroups of CRC patients (Figure 2A,B). Figure 3 further illustrates how a GCC mRNA level over 300 copies/µL was stable and significantly correlated with reduced OS in CRC patients.
Multivariate Cox regression analysis of GCC mRNA for DFS and OS
According to the previous data, the variables with P value <0.05 were selected based on univariate Cox regression model analysis and were analyzed via a multivariate Cox regression model method equivalent to Backward Stepwise (conditional LR) analysis. Multivariate Cox statistical survival analysis showed a significant correlation of (I) OS with GCC mRNA levels, tumor emboli in vessels, mesenteric root lymph node metastases, and differentiation types; and (II) DFS with GCC mRNA levels, emboli in vessels, mesenteric root lymph node metastases, and CA199 levels. The correlation between OS or DFS in the current study and the following six factors was evaluated via Kaplan-Meier survival curves: (I) tumor emboli in vessels, (II) mesenteric root lymph node metastases, (III) CA199 level in peripheral blood, (IV) differentiation type (see Figure S1A,B,C,D,E,F), and (V) circulating GCC mRNA level (see Figure S2A,B,C,D,E,F,G,H). The log-rank test showed significant differences between OS and DFS with respect to GCC mRNA levels over baseline, and other characteristics (all P<0.05).
A nomogram for predicting the survival of CRC patients
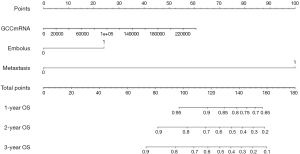
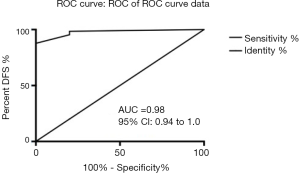
On the basis of the results above, it was revealed that the GCC mRNA copy number could serve as a potential metastatic predictor in the diagnosis of colorectal cancer. It was also observed that several clinical parameters including tumor embolus in vessels and metastases, were associated with OS. Accordingly, we built a nomogram based on both molecular biomarkers and clinical variables (Figure 4). For 90 patients who were randomly selected, points were assigned to the following three variables: tumor embolus in vessel, metastasis, and GCC mRNA copy number. From this, a total score was calculated from the nomogram. The total scores corresponded to survival predictions for 1/2/3-year OS. The nomogram was further validated internally by bootstrap resampling (n=70). The survival prediction obtained from the bootstrap correction and the actual survival is shown in the calibration plot with the area under the curve (AUC) 0.98 (Figure 5).
Discussion
The detection of CTCs in the peripheral blood is selectively used for metastatic colorectal cancer patients in the prediction of distant organ metastasis and the assessment of clinical treatment or long-term survival (4-6). However, for early-stage CRC patients, the qRT-PCR-based technique is commonly applied for the detection of CTCs and is thus dependent on specific biomarkers and sensitive techniques (8,9,12). Although epithelial-related biomarkers have been widely used in recent diagnosis, treatment, and prognosis of CRC patients, there remains a lack of a specific marker that is stably expressed in CRCs during the epithelial-mesenchymal transition (EMT) process (26,27). GCC is one of the most specific and sensitive biomarkers that can be stably expressed in intestinal tumor cells from primary tissue sources to metastatic organs (28-30). GCC is over-expressed selectively in human colorectal carcinoma, but not in extra-intestinal tissues and tumors, indicating its possibility as a specific CRC biomarker for CTC detection and capture (17). Furthermore, the GCC mRNA-based CTC search method might increase the detection compared to the other tumor markers in metastasized CRC cells (13,31). GCC has been reported as a suitable molecular biomarker for micro-metastasis in lymph nodes, periphery blood, and metastatic organs of CRC patients (32-34), while few studies have explored the potential correlation of GCC mRNA with OS or DFS. In this study, the internal correlation of GCC mRNA with patients’ metastasis and survival was investigated, and the clinical value of GCC mRNA as metastasis and prognostic marker in CRC patients was revealed.
The present studies and trials have highlighted the enumeration of CTCs as an important biomarker for survival prediction and for the therapeutic monitoring of metastatic CRC patients (22-24). It was reported that CTC amounts below or over the cutoff value (about 5 CTCs) indicated negative or positive results of current treatment in metastatic cases, but few studies explored the correlation between the cutoff value of circulating mRNA and DFS or OS in early-stage CRC patients (8,23,31). In this study, GCC mRNA was revealed by statistical analysis to be a prognostic biomarker in CRC patients. This is the first study to establish the significant correlation of reduced OS with GCC mRNA and reduced DFS with GCC mRNA (>100 copies/µL or higher than median value). Then, based on stage stratification, reduction of OS and DFS were found to be significantly associated with high GCC mRNA copy number in the stage II subgroup. According to UICC staging, the stage II subgroup presented with muscular and vein invasion without lymph node or distal organ metastasis, and thus, chemotherapy was required after surgical treatment with the potential hazard of hematogenous spread of tumor cells (24). The accurate selection of stage II subgroup with the hazard of tumor spread and metastasis for chemotherapy remains challenging. Herein, it was indicated that the level of GCC mRNA in circulation is a valuable index for the selection of stage II CRC patients with high hazard due to its strong correlation with DFS and OS (13,33). In addition, the correlation of a reduction of DFS with GCC mRNA level >8,000 copies/µL and a reduction of OS with GCC mRNA level >1,500 copies/µL in stage I subgroups was apparent, which indicated that early stage cases were also prone to tumor spread and potential hazard of distant organ metastasis (27). Therefore, for the stage I and II CRC patients with high GCC mRNA levels, routine examination, immunotherapy, and chemotherapy were required after surgical treatment.
Furthermore, a stable and significant correlation was observed between the GCC mRNA level higher than >300 copies/µL and OS decrease in CRC patients, which was consistent with the cutoff value calculated by the distribution curve. Based on the above statistical analysis, as the cutoff value in the early stage was higher than that in advanced stages, the cutoff value of GCC mRNA should be established according to the specific clinical stage of a given CRC patient. In this study, the suitable cutoff value for stage II CRC patients was 100 copies/µL, while that for stage I was 1,500 copies/µL or more, which was different from that in metastatic cases. Consequently, a nomogram was set up based on variables including tumor embolus in vessel, metastasis, and GCC mRNA copy number, which showed that the total score corresponded closely with the survival prediction for 1/2/3-year OS. Therefore, our study strongly demonstrates that GCC mRNA can be a reliable circulating biomarker mRNA for survival prediction and tumor metastasis in CRC patients, and can be used in the clinical application of GCC in the future.
In conclusion, circulating GCC mRNA was shown to be a reliable metastasis predictor and prognostic marker in early stage CRC patients, and can thus provide guidance in the early clinical treatment before tumor spread. However, large sample trials are still required in the future to confirm GCC mRNA as metastasis and prognostic biomarker in early-stage CRC patients.
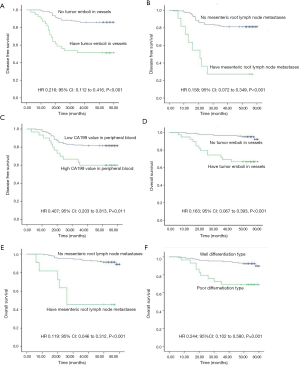
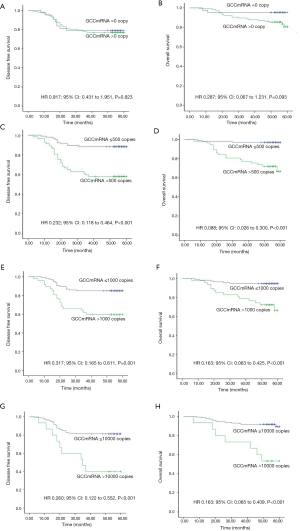
Table S1
| Variables | N (%) | 5-year DFS times (months) | 5-year OS times (months) | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| Age | ||||||
| ≤50 years | 45 (28.1) | Reference | Reference | |||
| >50 years | 115 (71.9) | 1.253 (0.627–2.506) | 0.523 | 0.773 (0.283–2.110) | 0.615 | |
| Sex | ||||||
| Females | 59 (36.9) | Reference | Reference | |||
| Males | 101 (63.1) | 0.936 (0.474–1.848) | 0.849 | 0.657 (0.255–1.695) | 0.386 | |
| Tumor location | ||||||
| Colon | 60 (37.5) | Reference | Reference | |||
| Rectum | 100 (62.5) | 1.272 (0.656–2.468) | 0.476 | 1.636 (0.693–3.867) | 0.262 | |
| Tumor size | ||||||
| Less than 5 cm | 108 (67.5) | Reference | Reference | |||
| More than 5 cm | 52 (32.5) | 0.942 (0.471–1.883) | 0.865 | 0.416 (0.177–0.981) | 0.045* | |
| Pathological type | ||||||
| Ulcerative type | 46 (28.8) | Reference | Reference | |||
| Protruded | 114 (71.3) | 2.170 (1.124–4.189) | 0.021* | 2.113 (0.888–5.028) | 0.091 | |
| Differentiation type | ||||||
| Well | 131 (81.9) | Reference | Reference | |||
| Poor | 29 (18.1) | 0.418 (0.206–0.850) | 0.016* | 0.244 (0.102–0.580) | 0.001* | |
| Infiltration depth | ||||||
| Mucosal and muscularis infiltrating | 40 (25.0) | Reference | Reference | |||
| Serosal invasion | 120 (75.0) | 0.819 (0.373–1.798) | 0.619 | 0.670 (0.225–1.991) | 0.471 | |
| Tumor emboli in vessels, | ||||||
| No | 121 (75.6) | Reference | Reference | |||
| Yes | 39 (24.4) | 0.216 (0.112–0.416) | 0.000* | 0.163 (0.067–0.393) | 0.000* | |
| Lymph node metastases | ||||||
| No | 90 (56.3) | Reference | Reference | |||
| Yes | 70 (43.8) | 0.299 (0.147–0.607) | 0.001* | 0.360 (0.145–0.893) | 0.027* | |
| Mesenteric root lymph node metastases | ||||||
| No | 149 (93.1) | Reference | Reference | |||
| Yes | 11 (6.9) | 0.158 (0.072–0.349) | 0.000* | 0.119 (0.046–0.312) | 0.000* | |
| TNM stage | ||||||
| Stage I | 31 (19.4) | Reference | Reference | |||
| Stage II | 58 (36.3) | 0.403 (0.154–1.054) | 0.064 | 0.477 (0.128–1.560) | 0.207 | |
| Stage III | 71 (44.4) | 0.257 (0.105–0.626) | 0.003* | 0.331 (0.109–1.005) | 0.051 | |
| CA199 values in peripheral blood | ||||||
| ≤37 U/dL | 130 (81.3) | Reference | Reference | |||
| >37 U/dL | 30 (18.8) | 0.407 (0.203–0.813) | 0.011* | 0.531 (0.206–1.370) | 0.191 | |
| CEA values in peripheral blood | ||||||
| ≤5 ng/mL | 102 (63.8) | Reference | Reference | |||
| >5 ng/mL | 58 (36.3) | 0.538 (0.280–1.033) | 0.063 | 0.551 (0.233–1.307) | 0.176 | |
| Postoperative chemotherapy | ||||||
| No | 59 (36.9) | Reference | Reference | |||
| Venous chemotherapy | 64 (40.0) | 0.899 (0.320–2.526) | 0.840 | 1.088 (0.318–3.722) | 0.893 | |
| Oral chemotherapy | 37 (23.1) | 2.210 (0.892–5.476) | 0.087 | 1.579 (0.495–5.037) | 0.440 | |
Results were represented as hazard ratio (HR) with respective 95% confidence interval of HR (95% CI) through univariate Cox-regression model analysis. *, P<0.05, indicates significant. DFS, disease-free survival; OS, overall survival; CEA, carcinoembryonic antigen.
Table S2
| mRNA copy | GCC, n (%) | GCC, DFS | GCC, OS | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| 0 | ||||||
| =0 | 43 (26.9) | Reference | Reference | |||
| >0 | 117 (73.1) | 0.917 (0.431–1.951) | 0.823 | 0.287 (0.067–1.231) | 0.093 | |
| 50 | ||||||
| ≤50 | 59 (36.9) | Reference | Reference | |||
| >50 | 101 (63.1) | 0.547 (0.257–1.163) | 0.117 | 0.184 (0.043–0.791) | 0.023* | |
| 100 | ||||||
| ≤100 | 74 (46.3) | Reference | Reference | |||
| >100 | 86 (53.8) | 0.355 (0.167–0.756) | 0.007* | 0.122 (0.028–0.526) | 0.005* | |
| 200 | ||||||
| ≤200 | 84 (52.5) | Reference | Reference | |||
| >200 | 76 (47.5) | 0.268 (0.126–0.570) | 0.001* | 0.093 (0.022–0.403) | 0.001* | |
| 300 | ||||||
| ≤300 | 95 (59.4) | Reference | Reference | |||
| >300 | 65 (40.6) | 0.225 (0.108–0.466) | 0.000* | 0.069 (0.016–0.298) | 0.000* | |
| 400 | ||||||
| ≤400 | 103 (64.4) | Reference | Reference | |||
| >400 | 57 (35.6) | 0.232 (0.118–0.464) | 0.000* | 0.088 (0.026–0.300) | 0.000* | |
| 500 | ||||||
| ≤500 | 103 (64.4) | Reference | Reference | |||
| >500 | 57 (35.6) | 0.232 (0.118–0.464) | 0.000* | 0.088 (0.026–0.300) | 0.000* | |
| 600 | ||||||
| ≤600 | 104 (65.0) | Reference | Reference | |||
| >600 | 56 (35.0) | 0.224 (0.112–0.449) | 0.000* | 0.085 (0.025–0.289) | 0.000* | |
| 700 | ||||||
| ≤700 | 109 (68.1) | Reference | Reference | |||
| >700 | 51 (31.9) | 0.284 (0.146–0.551) | 0.000* | 0.142 (0.052–0.391) | 0.000* | |
| 800 | ||||||
| ≤800 | 110 (68.8) | Reference | Reference | |||
| >800 | 50 (31.3) | 0.274 (0.141–0.532) | 0.000* | 0.138 (0.050–0.380) | 0.000* | |
| 900 | ||||||
| ≤900 | 110 (68.8) | Reference | Reference | |||
| >900 | 50 (31.3) | 0.274 (0.141–0.532) | 0.000* | 0.138 (0.050–0.380) | 0.000* | |
| 1,000 | ||||||
| ≤1,000 | 113 (70.6) | Reference | Reference | |||
| >1,000 | 47 (29.4) | 0.317 (0.165–0.611) | 0.001* | 0.163 (0.063–0.425) | 0.000* | |
| 1,500 | ||||||
| ≤1,500 | 123 (76.9) | Reference | Reference | |||
| >1,500 | 37 (23.1) | 0.427 (0.218–0.835) | 0.013* | 0.225 (0.093–0.544) | 0.001* | |
| 2,000 | ||||||
| ≤2,000 | 125 (78.1) | Reference | Reference | |||
| >2,000 | 35 (21.9) | 0.392 (0.200–0.766) | 0.006* | 0.207 (0.086–0.500) | 0.000* | |
| 2,500 | ||||||
| ≤2,500 | 132 (82.5) | Reference | Reference | |||
| >2,500 | 28 (17.5) | 0.372 (0.186–0.744) | 0.005* | 0.188 (0.078–0.451) | 0.000* | |
| 3,000 | ||||||
| ≤3,000 | 135 (84.4) | Reference | Reference | |||
| >3,000 | 25 (15.6) | 0.378 (0.186–0.768) | 0.007* | 0.202 (0.084–0.487) | 0.000* | |
| 3,500 | ||||||
| ≤3,500 | 138 (86.3) | Reference | Reference | |||
| >3,500 | 22 (13.8) | 0.314 (0.154–0.639) | 0.001* | 0.169 (0.070–0.408) | 0.000* | |
| 4,000 | ||||||
| ≤4,000 | 140 (87.5) | Reference | Reference | |||
| >4,000 | 20 (12.5) | 0.323 (0.156–0.670) | 0.002* | 0.183 (0.075–0.448) | 0.000* | |
| 4,500 | ||||||
| ≤4,500 | 140 (87.5) | Reference | Reference | |||
| >4,500 | 20 (12.5) | 0.323 (0.156–0.670) | 0.002* | 0.192 (0.079–0.464) | 0.000* | |
| 5,000 | ||||||
| ≤5,000 | 141 (88.1) | Reference | Reference | |||
| >5,000 | 19 (11.9) | 0.362 (0.170–0.771) | 0.008* | 0.223 (0.089–0.560) | 0.001* | |
| 6,000 | ||||||
| ≤6,000 | 142 (88.8) | Reference | Reference | |||
| >6,000 | 18 (11.3) | 0.336 (0.158–0.715) | 0.005* | 0.208 (0.083–0.523) | 0.001* | |
| 7,000 | ||||||
| ≤7,000 | 144 (90.0) | Reference | Reference | |||
| >7,000 | 16 (10.0) | 0.285 (0.134–0.606) | 0.001* | 0.177 (0.070–0.444) | 0.000* | |
| 8,000 | ||||||
| ≤8,000 | 145 (90.6) | Reference | Reference | |||
| >8,000 | 15 (9.4) | 0.260 (0.122–0.552) | 0.000* | 0.163 (0.065–0.409) | 0.000* | |
| 9,000 | ||||||
| ≤9,000 | 145 (90.6) | Reference | Reference | |||
| >9,000 | 15 (9.4) | 0.260 (0.122–0.552) | 0.000* | 0.163 (0.065–0.409) | 0.000* | |
| 10,000 | ||||||
| ≤10,000 | 145 (90.6) | Reference | Reference | |||
| >10,000 | 15 (9.4) | 0.260 (0.122–0.552) | 0.000* | 0.163 (0.065–0.409) | 0.000* | |
Results were represented as hazard ratio (HR) with respective 95% confidence interval of HR (95% CI) through univariate Cox-regression model analysis. *, P<0.05, indicates significant. GCC, guanylyl cyclase C; DFS, disease-free survival; OS, overall survival.
Table S3
| mRNA copy | GCC, DFS (P value) | GCC, OS (P value) | |||||
|---|---|---|---|---|---|---|---|
| Stage I | Stage II | Stage III | Stage I | Stage II | Stage III | ||
| 0 | 0.481 | 0.102 | 0.533 | 0.829 | 0.215 | 0.137 | |
| 50 | 0.757 | 0.055 | 0.588 | 0.616 | 0.186 | 0.040* | |
| 100 | 0.536 | 0.017* | 0.085 | 0.463 | 0.100 | 0.006* | |
| 200 | 0.343 | 0.003* | 0.039* | 0.323 | 0.042* | 0.003* | |
| 300 | 0.343 | 0.000* | 0.011* | 0.323 | 0.011* | 0.000* | |
| 400 | 0.259 | 0.000* | 0.008* | 0.260 | 0.002* | 0.001* | |
| 500 | 0.259 | 0.000* | 0.008* | 0.260 | 0.002* | 0.001* | |
| 600 | 0.259 | 0.000* | 0.004* | 0.260 | 0.002* | 0.000* | |
| 700 | 0.483 | 0.000* | 0.030* | 0.102 | 0.002* | 0.010* | |
| 800 | 0.483 | 0.000* | 0.030* | 0.102 | 0.001* | 0.010* | |
| 900 | 0.483 | 0.000* | 0.030* | 0.102 | 0.001* | 0.010* | |
| 1,000 | 0.483 | 0.000* | 0.115 | 0.102 | 0.001* | 0.024* | |
| 1,500 | 0.240 | 0.032* | 0.324 | 0.034* | 0.022* | 0.062* | |
| 2,000 | 0.240 | 0.032* | 0.152 | 0.034* | 0.022* | 0.025* | |
| 2,500 | 0.240 | 0.001* | 0.337 | 0.034* | 0.001* | 0.042* | |
| 3,000 | 0.138 | 0.001* | 0.481 | 0.014* | 0.001* | 0.106 | |
| 3,500 | 0.138 | 0.001* | 0.119 | 0.014* | 0.001* | 0.020* | |
| 4,000 | 0.138 | 0.029* | 0.055 | 0.014* | 0.112 | 0.007* | |
| 4,500 | 0.138 | 0.029* | 0.055 | 0.014* | 0.112 | 0.007* | |
| 5,000 | 0.138 | 0.029* | 0.151 | 0.014* | 0.112 | 0.053 | |
| 6,000 | 0.138 | 0.009* | 0.151 | 0.014* | 0.057 | 0.053 | |
| 7,000 | 0.060 | 0.001* | 0.151 | 0.004* | 0.019* | 0.053 | |
| 8,000 | 0.015* | 0.001* | 0.151 | 0.000* | 0.019* | 0.053 | |
| 9,000 | 0.015* | 0.001* | 0.151 | 0.000* | 0.019* | 0.053 | |
| 10,000 | 0.015* | 0.001* | 0.151 | 0.000* | 0.019* | 0.053 | |
*, P<0.05, indicates statistically significant. GCC, guanylyl cyclase C; DFS, disease-free survival; OS, overall survival.
Acknowledgments
We thank all the patients treated at the Surgical Department of Colorectal Cancer of Zhejiang Cancer Hospital, who agreed to provide samples for this study.
Funding: This work was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.34). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Zhejiang Cancer Hospital (No. IRB-2019-171) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Riggi N, Aguet M, Stamenkovic I. Cancer Metastasis: A Reappraisal of Its Underlying Mechanisms and Their Relevance to Treatment. Annu Rev Pathol 2018;13:117-40. [Crossref] [PubMed]
- Chambers AF, Werb Z. Invasion and metastasis--recent advances and future challenges. J Mol Med (Berl) 2015;93:361-8. [Crossref] [PubMed]
- Chou WC, Wu MH, Chang PH, et al. A Prognostic Model Based on Circulating Tumour Cells is Useful for Identifying the Poorest Survival Outcome in Patients with Metastatic Colorectal Cancer. Int J Biol Sci 2018;14:137-46. [Crossref] [PubMed]
- Yang C, Zou K, Zheng L, et al. Prognostic and clinicopathological significance of circulating tumor cells detected by RT-PCR in non-metastatic colorectal cancer: a meta-analysis and systematic review. BMC Cancer 2017;17:725. [Crossref] [PubMed]
- Wang L, Zhou S, Zhang W, et al. Circulating tumor cells as an independent prognostic factor in advanced colorectal cancer: a retrospective study in 121 patients. Int J Colorectal Dis 2019;34:589-97. [Crossref] [PubMed]
- Messaritakis I, Sfakianaki M, Papadaki C, et al. Prognostic significance of CEACAM5mRNA-positive circulating tumor cells in patients with metastatic colorectal cancer. Cancer Chemother Pharmacol 2018;82:767-75. [Crossref] [PubMed]
- Tol J, Koopman M, Miller MC, et al. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol 2010;21:1006-12. [Crossref] [PubMed]
- Rahbari NN, Aigner M, Thorlund K, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology 2010;138:1714-26. [Crossref] [PubMed]
- Lapin M, Tjensvoll K, Oltedal S, et al. MINDEC-An Enhanced Negative Depletion Strategy for Circulating Tumour Cell Enrichment. Sci Rep 2016;6:28929. [Crossref] [PubMed]
- Lu Y, Liang H, Yu T, et al. Isolation and characterization of living circulating tumor cells in patients by immunomagnetic negative enrichment coupled with flow cytometry. Cancer 2015;121:3036-45. [Crossref] [PubMed]
- Thorsteinsson M, Jess P. The clinical significance of circulating tumor cells in non-metastatic colorectal cancer--a review. Eur J Surg Oncol 2011;37:459-65. [Crossref] [PubMed]
- Mohammadi P, Saidijam M, Kaki A, et al. A Pilot Study of CK19, CK20 and GCC mRNA in the Peripheral Blood as a Colorectal Cancer Biomarker Panel. Int J Mol Cell Med 2016;5:30-6. [PubMed]
- Blomain ES, Merlino DJ, Pattison AM, et al. Guanylyl Cyclase C Hormone Axis at the Intersection of Obesity and Colorectal Cancer. Mol Pharmacol 2016;90:199-204. [Crossref] [PubMed]
- Pattison AM, Merlino DJ, Blomain ES, et al. Guanylyl cyclase C signaling axis and colon cancer prevention. World J Gastroenterol 2016;22:8070-7. [Crossref] [PubMed]
- Ahsan MK, Tchernychev B, Kessler MM, et al. Linaclotide activates guanylate cyclase-C/cGMP/protein kinase-II-dependent trafficking of CFTR in the intestine. Physiol Rep 2017; [Crossref] [PubMed]
- Ikpa PT, Sleddens HF, Steinbrecher KA, et al. Guanylin and uroguanylin are produced by mouse intestinal epithelial cells of columnar and secretory lineage. Histochem Cell Biol 2016;146:445-55. [Crossref] [PubMed]
- Brenna Ø, Bruland T, Furnes MW, et al. The guanylate cyclase C signaling pathway is down-regulated in inflammatory bowel disease. Scand J Gastroenterol 2015;50:1241-52. [Crossref] [PubMed]
- Fava TA, Desnoyers R, Schulz S, et al. Ectopic expression of guanylyl cyclase C in CD34+ progenitor cells in peripheral blood. J Clin Oncol 2001;19:3951-9. [Crossref] [PubMed]
- Blomain ES, Lin JE, Kraft CL, et al. Translating colorectal cancer prevention through the guanylyl cyclase C signaling axis. Expert Rev Clin Pharmacol 2013;6:557-64. [Crossref] [PubMed]
- Brudvik KW, Seeberg LT, Hugenschmidt H, et al. Detection of Circulating Tumor Cells at Surgery and at Follow-Up Assessment to Predict Survival After Two-Stage Liver Resection of Colorectal Liver Metastases. Ann Surg Oncol 2015;22:4029-37. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol 2009;20:1223-9. [Crossref] [PubMed]
- Sawada T, Araki J, Yamashita T, et al. Prognostic Impact of Circulating Tumor Cell Detected Using a Novel Fluidic Cell Microarray Chip System in Patients with Breast Cancer. EBioMedicine 2016;11:173-82. [Crossref] [PubMed]
- Sotelo MJ, Sastre J, Maestro ML, et al. Role of circulating tumor cells as prognostic marker in resected stage III colorectal cancer. Ann Oncol 2015;26:535-41. [Crossref] [PubMed]
- Liu Y, Qian J, Feng JG, et al. Detection of circulating tumor cells in peripheral blood of colorectal cancer patients without distant organ metastases. Cell Oncol (Dordr) 2013;36:43-53. [Crossref] [PubMed]
- Das V, Bhattacharya S, Chikkaputtaiah C, et al. The basics of epithelial-mesenchymal transition (EMT): A study from a structure, dynamics, and functional perspective. J Cell Physiol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Yörüker EE, Holdenrieder S, Gezer U. Blood-based biomarkers for diagnosis, prognosis and treatment of colorectal cancer. Clin Chim Acta 2016;455:26-32. [Crossref] [PubMed]
- Brenna Ø, Bruland T, Furnes MW, et al. Gustafsson BI: The guanylate cyclase-C signaling pathway is down-regulated in inflammatory bowel disease. Scand J Gastroenterol 2015;50:1241-52. [Crossref] [PubMed]
- Rappaport JA, Waldman SA. The Guanylate Cyclase C-cGMP Signaling Axis Opposes Intestinal Epithelial Injury and Neoplasia. Front Oncol 2018;8:299. [Crossref] [PubMed]
- Li P, Wuthrick E, Rappaport JA, et al. GUCY2C Signaling Opposes the Acute Radiation-Induced GI Syndrome. Cancer Res 2017;77:5095-106. [Crossref] [PubMed]
- Gazzaniga P, Raimondi C, Gradilone A, et al. Circulating tumor cells in metastatic colorectal cancer: do we need an alternative cutoff? J Cancer Res Clin Oncol 2013;139:1411-6. [Crossref] [PubMed]
- Basu N, Saha S, Khan I, et al. Intestinal cell proliferation and senescence are regulated by receptor guanylyl cyclase C and p21. J Biol Chem 2014;289:581-93. [Crossref] [PubMed]
- Hyslop T, Waldman SA. Guanylyl cyclase C as a biomarker in colorectal cancer. Biomark Med 2013;7:159-67. [Crossref] [PubMed]
- Gallery M, Zhang J, Bradley DP, et al. A monomethyl auristatin E-conjugated antibody to guanylyl cyclase C is cytotoxic to target-expressing cells in vitro and in vivo. PLoS One 2018;13:e0191046. [Crossref] [PubMed]


