
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-22-2239 the article has been update on 2022-08-10 at here.
Silencing of DHX32 increases the proliferation of liver cancer cells
Introduction
Liver cancer ranks fifth in malignancy incidence throughout the world (1) and is the second leading cause of cancer-related death in China (2). Chronic hepatitis B or C infection and alcohol abuse have been identified to be the major risk factors for liver cancer development (3,4), which is characterized by its high complexity and heterogeneity.
RNA helicases, as motor proteins, modulate the structure of RNA by catalyzing the unwinding of duplex nucleic acids (5). DHX32, an RNA helicase, belongs to the DEAH box RNA helicase family by virtue of its homology to the conserved helicase domain (6). DHX32 is abundantly expressed in human and murine models (7), participating in cell transcription and differentiation, and being capable of regulating T-cell apoptosis (8). DHX32 has also been associated with some malignant tumors. Alli et al. found that DHX32 is dysregulated in certain types of lymphoma, suggesting its differential expression pattern in lymphoma (9). The mRNA level of DHX32 has been observed to be consistently high in colorectal cancer (CRC) tissues, and its level is correlated with tumor distribution, lymph node metastasis, nodal status, differentiated grade, and Dukes stage of CRC (10). DHX32 has also been found to enhance the proliferative, migratory, and invasive potential of CRC (11). Astuti et al. described the involvement of DHX32 in inherited retinal diseases (12), and it has also been reported to be upregulated in breast cancer tissues (13). Taken together, this evidence indicates that DHX32 plays a crucial role in tumor progression. The role of DHX32 in liver cancer, however, is unclear.
In view of the high incidence and mortality of liver cancer, it is necessary to explore the role of DHX32 in the development of this deadly disease. This study detected the expression of DHX32 in liver cancer tissues and their paracancerous tissues and analyzed the correlation between DHX32 expression and the pathology of liver cancer. In addition, HepG2 and Huh-7 cells with DHX32 knockdown were established by shRNA transfection to investigate the potential function of DHX32 in regulating behaviors of liver cancer cells.
Methods
Patients
Fifty-three liver cancer tissue and paracancerous tissue samples were surgically resected from 53 patients who were admitted to Zhongshan Hospital between 2006 and 2008. Tissue samples were preserved at the Open Laboratory Center of Zhongshan Hospital. All patients were pathologically diagnosed with primary liver cancer and did not receive anti-cancer therapy before tumor resection. All 53 patients had a hepatitis B infection. Imaging showed that all 53 cases of liver cancer were in situ, with no patients having distant metastasis. Informed consent was obtained prior to surgery, and study approval was given by the hospital ethics committee.
Tissue microarray (TMA) construction
Liver cancer TMAs were constructed using tissue cores from formalin-fixed, paraffin-embedded specimens, as previously described (14). A pathologist was responsible for reviewing the hematoxylin and eosin (H&E)-stained slides to determine tumor location. Representative tumor regions and their paracancerous tissues containing 2 tissue cores in each region were taken using an automated tissue arrayer (Beecher Instruments, Sun Prairie, WI, USA). Cores were transferred to individual recipient blocks. Sections (5 µm) were cut from each recipient block to confirm the presence of the tumor.
Immunohistochemistry (IHC)
IHC was constructed, as previously described (15). Primary polyclonal antibody DHX32 (dilution 1:100, rabbit, no. 19808-1-A, Proteintech, Chicago, IL, USA) was incubated overnight at 4 °C. The next day, slides were incubated with corresponding secondary biotinylated rabbit antibody (dilution 1:100, goat, no. A16100, Thermo Fisher Scientific Inc., MA, USA) for 30 min at 37 °C and washed with Tris-buffered saline. After incubation with strep-avidin-conjugated horseradish peroxidase (dilution 1:100, Dako, Glostrup, Denmark) for 30 min at 37 °C, chromogenic immunolocalization was conducted using 0.05% 3,3-diaminobenzidine tetrahydrochloride. Other cores containing liver cancer were used as positive controls. Non-immune rabbit serum (10%) was used as the negative control of the primary antibody. Sigma hematoxylin was used as a counterstain for IHC.
DHX32 expression was scored as the percentage of stained tumor cells in the section (0, none; 1, <20%; 2, 20–75%; 3, >75%). Staining intensity was graded from 0 to 3 (0, none; 1, weak; 2, intermediate; 3, strong). The total score was calculated as follows: score = intensity × positive rate. The expression level of DHX32 was categorized as low (≤6) or high (>6). A light microscope (Olympus Corporation, Tokyo, Japan) was used to observe the cells. The immunostained microarray sections were independently analyzed by two pathologists blinded to the clinicopathological information.
Cell culture
Liver cancer cell lines HepG2 and Huh-7 were cultured in Dulbecco’s modified Eagle’s medium (DMEM), containing 10% GibcoTM fetal bovine serum (FBS) and 100 IU/MI penicillin-streptomycin (Sigma-Aldrich/Merck KGaA, Darmstadt, Germany). HepG2 and Huh-7 cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA).
ShRNA synthesis and construction of Plvthm-DHX32-RNAi(s)/NC plasmids
The human shRNA sequences of DHX32 were designed using a BLOK-iT RNAi designer (Invitrogen, Carlsbad, CA, USA). The shRNA and control sequences (Table 1) were inserted into the MscI-Fsp target sequences of the Plv-mCherry-N (Invogen Tech. Co., Beijing, China) to generate Plv-mCherry-N-DHX32-RNAi(s) and Plv-mCherry-N -DHX32-NC plasmids.
Table 1
| Target gene | Selected sequence |
|---|---|
| ShDHX32-876 | 5'-CTATCAAGGATCTAACCTAAA-3' |
| ShDHX32-1898 | 5'-ATGGATCAGGTAACTACTTAA-3' |
| ShDHX32-1868 | 5'- AGTGGGTCCTCTTCCATAAAT-3' |
| Negative control (NC) | 5'- GTTCTCCGAACGTGTCACGT-3' |
Establishment of cells with stable DHX32 knockdown
HepG2 and Huh-7 cells were plated into 6-well plates (3×105 cells per well), 96-well plates (1×104 cells per well), or 12-well plates (1.5×105 cells per well) for 24 h before transfection. HepG2 and Huh-7 cells were transfected with Plv-mCherry-N-DHX32-RNAi(s) or Plv-mCherry-N-DHX32-NC using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). Neomycin-resistant colonies were subcultured with puromycin (9.0 µg/mL, Sigma-Aldrich/Merck KGaA, Darmstadt, Germany) for 14 days. The transfection efficacy of shRNA was verified by Western blotting. Cells with stable DHX32 knockdown were named HepG2-1868, HepG2-1898, Huh-7-1868, and Huh-7-1898 cells, and those transfected with controls were named HeG2-NC and Huh7-NC. These experiments were repeated in triplicate.
Western blot
The protein extraction and Western blot analysis were constructed as previously described (15). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. The primary antibodies of Akt (dilution 1:1,000, rabbit polyclonal, no.10176-2-AP), phosphorylated Akt (Ser473) [p-Akt (Ser473), dilution 1:3,000, mouse monoclonal, no.4051S] and ERK1/2 (dilution 1:1,000, rabbit polyclonal, no. 16443-1-AP) were purchased from Proteintech (Chicago, IL, USA). Phosphorylated ERK1/2 (Thr202/Tyr204) [p-ERK1/2 (Thr202/Tyr204), dilution 1:1,000, rabbit, no. 20G11] and CDK6 (dilution 1:2,000, mouse monoclonal, no. 3136) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). GAPDH antibody (dilution 1:2,000, rabbit, no.10493-1-AP) and the secondary antibodies horseradish-peroxidase (HRP)-conjugated goat anti-rabbit IgG (dilution 1:10,000; catalog no. AS014), and HRP-conjugated goat anti-mouse IgG (dilution 1:10,000; catalog no. AS003) were purchased from Abcam (Cambridge, MA, USA). The specifically bound antibodies were detected with enhanced chemiluminescence (ECL) (Millipore Co. Billerica, MA, USA). Images were analyzed using the BIO Photometer (Eppendorf AG, Hamburg, Germany).
Cell proliferation determination
For methyl thiazolyl tetrazolium (MTT) (Sigma, St. Louis, MO, USA) colorimetric assay, cells were seeded in 96-well plates with 2×103 cells/well. Next, 20 mL aliquot of MTT (5 mg/mL) was applied per well for 4-h incubation, followed by solubilization of formazan precipitate in 150 mL of dimethylsulfoxide (DMSO, Sigma, St. Louis, MO, USA). A microplate reader (MR-96A, Mindray, Shanghai, China) was used for recording the absorbance at 490 nm. For the EdU assay, cells were subjected to 25 µM 5-ethynyl-2'-deoxyuridine (EdU, RiboBio, Guangzhou, China) exposure for 2 h at 37 °C, with 4% PFA (RiboBio) fixation and 0.5% Triton-X (RiboBio) permeabilization. After incubation with 16 Apollo reaction cocktail (RiboBio) for 30 min, nuclei were subjected to 30-min dye with 4',6-diamidino-2-phenylindole (DAPI, Bepharm, Ltd. Shanghai, China) and captured under laser scanning confocal microscopy (TCS Sp5, Leica, Germany). All experiments were performed in triplicate.
Statistical analyses
Statistical Package for the Social Sciences (SPSS) v.13.0 (SPSS, Inc.) and GraphPad Prism 5 (GraphPad Software, Inc., CA, USA) were introduced for statistical analysis. The correlation between DHX32 expression and pathology in liver cancer patients was analyzed using the χ2 test. The differences between the two groups were analyzed by group Student’s t-test. Since only two patients were histologic grade I, we combined patients with histologic grade I and II together. P<0.05 indicated statistical significance.
Results
Baseline characteristics of enrolled liver cancer patients
Baseline characteristics of enrolled liver cancer patients are summarized in Table 2, including 42 males and 11 females with an average age of 53.5 years (ranging from 19 to 77 years). In terms of tumor stage, 3.8% of cases were stage I, 60.4% were stage II, and 35.8% were stage III. No patients experienced distant metastasis.
Table 2
| Characteristics | Value |
|---|---|
| Age (years) | |
| Median | 53.5 |
| Range | 19–77 |
| Sex, n (%) | |
| Female | 11 (20.8) |
| Male | 42 (79.2) |
| Histologic grade | |
| I | 2 (3.8) |
| II | 32 (60.4) |
| III | 19 (35.8) |
HCC, hepatocellular carcinoma.
Expression of DHX32 in liver cancer tissues and paracancerous tissues
To determine the distribution of DHX32 in 53 pairs of liver cancer tissues and the paracancerous tissues, positive expression of DHX32 was determined with IHC. Typical representative immunohistochemical results from one pair of hepatocellular carcinoma (HCC) tissue and adjacent noncancerous liver tissue using an anti-DHX32 antibody (Figure 1). Figure 1A shows that the positive expression of DHX32 was lower in liver cancer tissues, while there was strong cytoplasm staining of DHX32 in paracancerous tissues relative to liver cancer tissues (Figure 1B). The results of 53 pairs IHC showed that 88.7% (47/53) of paracancerous tissues exhibited a high expression (score >6) of DHX32, which was only 43.4% (23/53) in liver cancer tissues (Table 3). The IHC results suggest that DHX32 may play an important role in liver cancer development.
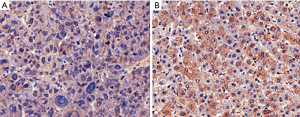
Table 3
| Tissues | N | DHX32 (protein) | ||
|---|---|---|---|---|
| Low* | High* | P value | ||
| The hepatocellular carcinoma tissues | 53 | 30 | 23 | <0.001 |
| The adjacent noncancerous tissues | 53 | 6 | 47 | |
*, DHX32 expression was scored as the percentage of stained tumor cells in the section (0, none; 1, <20%; 2, 20–75%; 3, >75%). Staining intensity was graded from 0 to 3 (0, none; 1, weak; 2, intermediate; 3, strong). Total score was calculated: Score = intensity × positive rate. Expression level of DHX32 was categorized as low (≤6) or high (>6).
We further elucidated the correlation between DHX32 abundance and pathological features of liver cancer. However, we did not observe any correlation between DHX32 expression and sex, age, tumor position, tumor size, or pathological grade of liver cancer patients (Table 4).
Table 4
| Variables | DHX32 expression | ||
|---|---|---|---|
| Low* | High* | P value** | |
| Age (years) | 0.328 | ||
| <60 | 20 | 16 | |
| ≥60 | 7 | 10 | |
| Gender | 0.990 | ||
| Female | 6 | 5 | |
| Male | 23 | 19 | |
| Tumor position | 0.415 | ||
| Left liver | 11 | 6 | |
| Right liver | 13 | 16 | |
| Left + right liver | 4 | 3 | |
| Histologic grade | 0.530 | ||
| I+II | 19 | 20 | |
| III | 11 | 3 | |
| Size | 0.743 | ||
| <4 cm | 8 | 7 | |
| ≥4 cm | 21 | 15 | |
*, DHX32 expression was scored as the percentage of stained tumor cells in the section (0, none; 1, <20%; 2, 20–75%; 3, >75%). Staining intensity was graded from 0 to 3 (0, none; 1, weak; 2, intermediate; 3, strong). Total score was calculated: score = intensity × positive rate. Expression level of DHX32 was categorized as low (≤6) or high (>6). **, P value of <0.05 is considered statistically significant.
Establishment of liver cancer cells with stable knockdown of DHX32
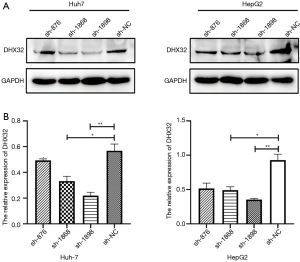
Cells were transfected with pLVTHM-DHX32-RNAi(s) or pLVTHM-DHX32-NC, respectively. Several clones from each transfection group were selected and analyzed by Western blot. DHX32-positive clones exhibited different levels of DHX32 expression in HepG2 and Huh-7 cells. HepG2-1868, HepG2-1898, Huh-7-1868, and Huh-7-1898 cells showed a relatively low level of DHX32, which were chosen for the subsequent experiments (Figure 2A,B). In addition, the transfection efficacy of sh-1898 was better than that of sh-1868. Liver cancer cell lines with stable silencing of DHX32 were named HepG2-siDHX32 and Huh-7-siDHX32.
Knockdown of DHX32 enhances the proliferative rate of liver cancer cells
The regulatory effect of DHX32 on the proliferative potential of HepG2-siDHX32 cells and Huh-7-siDHX32 cells was determined. MTT assay showed higher proliferative rates of HepG2-siDHX32 cells and Huh-7-siDHX32 cells relative to controls at 24, 48, 72, and 96 h (Figure 3A,B). Furthermore, the proliferative rate of HepG2-siDHX32 cells and Huh-7-siDHX32 cells transfected with sh-1898 were higher than those transfected with sh-1868 at 24, 72, and 96 h. However, we did not observe changes at the 48 h time point in Huh-7-siDHX32 cells.
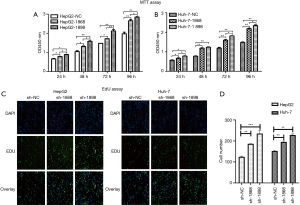
An EdU assay was conducted for revealing changes in cell growth. The percentage of EdU-positive cells increased in HepG2-siDHX32 cells and Huh-7-siDHX32 cells relative to the controls (Figure 3C,D). Both the MTT and EdU assays demonstrated that the DHX32 knockdown accelerated the proliferative rates of HepG2-siDHX32 and Huh-7-siDHX32 cells relative to controls.
DHX32 regulates the phosphorylation of Akt and ERK1/2 and also changes the expression of CDK6
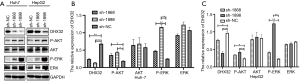
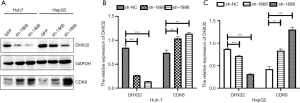
Previous studies have confirmed the crucial functions of Akt and ERK1/2 in cell proliferation and apoptosis (16,17). Here, expression levels of Akt and ERK1/2 were not influenced by DHX32. However, p-Akt and p-ERK1/2 were upregulated by the DHX32 knockdown (Figure 4A,B,C). DHX32 inhibited the phosphorylation of Akt and ERK1/2 in liver cancer cells. We also identified the effector that regulates the cell cycle progression of HepG2 and Huh7 cells by analyzing the expression of CDK6 (Figure 5A,B,C). The results showed that the expression of CDK6 was increased in DHX32 knockdown cells.
Discussion
In this study, we first observed that DHX32 had lower expression in liver cancer tissues than in their adjacent normal tissues. Meanwhile, MTT and EdU data demonstrated that DHX32 knockdown in liver cancer cells enhanced the proliferative potential of liver cancer cells. Furthermore, phosphorylated levels of ERK and Akt were upregulated in liver cancer cells with DHX32 knockdown. We also found the level of CDK6 increased in liver cancer cells with DHX32 knockdown. Together, these results support the notion that DHX32 plays a negative role in liver cancer proliferation.
Some dysregulated RNA helicases have already been found in various types of cancer. DHX9 persists at a high level in lung tumors compared to normal lung tissues (18). DDX6 is overexpressed in CRC and participates in the protein translation of c-myc by modifying the mRNA structure (19). Both mRNA and protein expressions of DDX3 are down-regulated in liver cancer specimens, while DDX3 may harbor a growth-suppressive ability (20). In this paper, IHC results revealed a decreased positive expression of DHX32 in liver cancer tissues relative to their paracancerous tissues. This thus suggests that DHX32 may contribute to the development of liver cancer.
Previous research has established that DHX32 participates in transcription and differentiation, regulation of critical immune response genes, and Fas-mediated apoptosis in T-cells (8,21,22). Lin et al. found that DHX32 enhances proliferative, migratory, and invasive potentials, but attenuates the chemotherapeutic sensitivity of CRC cells (11). Recently, Lin et al. reported that DHX32 upregulates the expression of vascular endothelial growth factor A (VEGFA) at the transcription level through interacting and stabilizing beta-catenin in CRC cells (23).
Despite these findings, the precise role of DHX32 in liver cancer is currently unknown. Through the MTT and EdU assays conducted in this study, we demonstrated the inhibitory effect of DHX32 on the proliferation of liver cancer cells, but there are other RNA helicases that have been implicated in cancer proliferation. For instance, DDX46 knockdown reduces the proliferative capacity of colorectal cancer (24), while DDX5 mediates the proliferation of lung cancer cells by activating the beta-catenin pathway (25). Meanwhile, Fidaleo et al. found that alternative pre-mRNA processing of DHX9 could cause genotoxic stress, thus inhibiting Ewing sarcoma cell growth (26). Here, it is particularly worth noting that DEAD helicase DDX3 inhibited the proliferation of liver cancer cells through upregulating p21waf1/cip1 expression (20) .
Akt and ERK are greatly involved in mediating proliferation (16,17), and the Akt pathway has been identified to be activated in liver cancer (27). Liu et al. also reported sorafenib-induced proliferative inhibition and apoptosis acceleration in liver cancer cells via MEK/ERK-dependent or independent pathways (28). Meanwhile, Liu et al. found that TTK promotes the proliferative and migratory abilities of liver cancer cells by activating the Akt/mTOR pathway (29). Schmitz et al. found that p-ERK1/2 and p-Akt were upregulated in liver cancer tissues, and activated ERK and Akt pathways could predict the unsatisfactory outcome of liver cancer (30). Finally, Chen et al. showed that epithelial cell transforming sequence 2 (ECT2) upregulation is correlated with the early recurrent and poor survival of liver cancer through activating Rho/ERK axis (31). Our own results showed that silencing DHX32 increased expressions of p-Akt and p-ERK in HepG2-siDHX32 and Huh-7-siDHX32 cells, providing evidence that DHX32 may inhibit proliferation of liver cancer cells through downregulating p-Akt and p-ERK expression in liver cancer cells.
The overexpression of CDK6, a cell-cycle kinase, has been detected in human liver cancer tissues (32) and has been shown to be directly involved in transcription in tumor cells and the regulation of their proliferation (33). Our Western blot analysis showed that knockdown of DHX32 induced the upregulation of CDK6 in liver cancer cells. This result further proves the inhibitory effect of DHX32 on the proliferation of liver cancer cells. As previous studies have shown that the PI3K/Akt and MEK/ERK pathways mediate the cell cycle (34,35), it would be interesting to determine whether CDK6 would be regulated by Akt or ERK-dependent pathways in HepG2 cells and Huh 7 cells in which DHX32 has been knocked down.
DHX32 is located on 10q26 chromosomal band and has 12 exons, with 2 of these exons having different transcripts regulated by two different promoters (9). The use of alternative promoters is an established mechanism in regulating tissue-specific and/or developmental stage-specific gene expression (36). This might explain the different distributions of DHX32 in colorectal cancer, breast cancer, some types of lymphoma and liver cancer, while also accounting for the opposite effect in colorectal cancer and liver cancer. In the future, we could determine the expression of the exon 1 transcript and exon 2 transcripts in liver cancer tissues and the paracancerous tissues to thus clarify the mechanism behind how DHX32 regulates tissue-specific and developmental stage-specific gene expression in liver cancer.
Some limitations of this study should also be addressed. Firstly, the expression pattern of DHX32 at mRNA and protein levels in liver cancer and the paracancerous tissues is still required. Secondly, the specific mechanism underlying DHX32 in regulating p-Akt and p-ERK has not been extensively clarified. Lastly, in vivo experiments are necessary to elucidate the proliferation-suppressing role of DHX32 in liver cancer further.
Conclusions
In conclusion, DHX32 showed low expression in liver cancer tissues compared to the paracancerous tissues and could harbor a proliferation-suppressing property in liver cancer. DHX32 may thus be utilized as a possible target for gene therapy.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.35). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The collection of liver cancer and paracancerous tissues in the present study was approved by the Medical Ethics Committee of Zhongshan Hospital, School of Medicine, Xiamen University, Xiamen (xmzsyyky-2018026). Informed consent was obtained prior to surgery.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang Y, Guo J, Hao Y, et al. Silencing of karyopherin alpha2 inhibits cell growth and survival in human hepatocellular carcinoma. Oncotarget 2017;8:36289-304. [PubMed]
- Li J, Cheng ZJ, Liu Y, et al. Serum thioredoxin is a diagnostic marker for hepatocellular carcinoma. Oncotarget 2015;6:9551-63. [PubMed]
- Wang W, Huang P, Wu P, et al. BCL6B expression in hepatocellular carcinoma and its efficacy in the inhibition of liver damage and fibrogenesis. Oncotarget 2015;6:20252-65. [PubMed]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118-27. [Crossref] [PubMed]
- Umate P, Tuteja N, Tuteja R. Genome-wide comprehensive analysis of human helicases. Commun Integr Biol 2011;4:118-37. [Crossref] [PubMed]
- Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics 2003;81:618-22. [Crossref] [PubMed]
- Alli Z, Ho M, Ackerley C, et al. Characterization of murine Dhx32. Exp Mol Pathol 2007;83:115-8. [Crossref] [PubMed]
- Alli Z, Chen Y, Abdul Wajid S, et al. A role for DHX32 in regulating T-cell apoptosis. Anticancer Res 2007;27:373-7. [PubMed]
- Alli Z, Ho M, Abdelhaleem M. Expression of DHX32 in lymphoid tissues. Exp Mol Pathol 2005;79:219-23. [Crossref] [PubMed]
- Huang C, Liang X, Huang R, et al. Up-regulation and clinical relevance of novel helicase homologue DHX32 in colorectal cancer. J Exp Clin Cancer Res 2009;28:11. [Crossref] [PubMed]
- Lin H, Liu W, Fang Z, et al. Overexpression of DHX32 contributes to the growth and metastasis of colorectal cancer. Sci Rep 2015;5:9247. [Crossref] [PubMed]
- Astuti GDN, van den Born LI, Khan MI, et al. Identification of Inherited Retinal Disease-Associated Genetic Variants in 11 Candidate Genes. Genes (Basel) 2018; [Crossref] [PubMed]
- Wang M, Zhang G, Wang Y, et al. DHX32 expression is an indicator of poor breast cancer prognosis. Oncol Lett 2017;13:942-8. [Crossref] [PubMed]
- He C, Jiang H, Geng S, et al. Expression of c-Myc and Fas correlates with perineural invasion of pancreatic cancer. Int J Clin Exp Pathol 2012;5:339-46. [PubMed]
- Wang JJ, Fang ZX, Ye HM, et al. Clinical significance of overexpressed cyclin-dependent kinase subunits 1 and 2 in esophageal carcinoma. Dis Esophagus 2013;26:729-36. [PubMed]
- Zhang W, Neo SP, Gunaratne J, et al. Feedback regulation on PTEN/AKT pathway by the ER stress kinase PERK mediated by interaction with the Vault complex. Cell Signal 2015;27:436-42. [Crossref] [PubMed]
- Sun Y, Liu WZ, Liu T, et al. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res 2015;35:600-4. [Crossref] [PubMed]
- Wei X, Pacyna-Gengelbach M, Schlüns K, et al. Analysis of the RNA helicase A gene in human lung cancer. Oncol Rep 2004;11:253-8. [PubMed]
- Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta 2004;1704:37-46.
- Chao CH, Chen CM, Cheng PL, et al. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 2006;66:6579-88. [Crossref] [PubMed]
- Alli Z, Nam EH, Beimnet K, et al. The activation-induced expression of DHX32 in Jurkat T cells is specific and involves calcium and nuclear factor of activated T cells. Cell Immunol 2005;237:141-6. [Crossref] [PubMed]
- Abdelhaleem M, Sun TH, Ho M. DHX32 expression suggests a role in lymphocyte differentiation. Anticancer Res 2005;25:2645-8. [PubMed]
- Lin H, Fang Z, Su Y, et al. DHX32 Promotes Angiogenesis in Colorectal Cancer Through Augmenting beta-catenin Signaling to Induce Expression of VEGFA. EBioMedicine 2017;18:62-72. [Crossref] [PubMed]
- Li M, Ma Y, Huang P, et al. Lentiviral DDX46 knockdown inhibits growth and induces apoptosis in human colorectal cancer cells. Gene 2015;560:237-44. [Crossref] [PubMed]
- Wang Z, Luo Z, Zhou L, et al. DDX5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating β-catenin signaling pathway. Cancer Sci 2015;106:1303-12. [Crossref] [PubMed]
- Fidaleo M, Svetoni F, Volpe E, et al. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget 2015;6:31740-57. [Crossref] [PubMed]
- Zender L, Villanueva A, Tovar V, et al. Cancer gene discovery in hepatocellular carcinoma. J Hepatol 2010;52:921-9. [Crossref] [PubMed]
- Liu L, Cao Y, Chen C, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res 2006;66:11851-8. [Crossref] [PubMed]
- Liu X, Liao W, Yuan Q, et al. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget 2015;6:34309-20. [PubMed]
- Schmitz KJ, Wohlschlaeger J, Lang H, et al. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol 2008;48:83-90. [Crossref] [PubMed]
- Chen J, Xia H, Zhang X, et al. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol 2015;62:1287-95. [Crossref] [PubMed]
- Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 2018;18:116. [Crossref] [PubMed]
- Tigan AS, Bellutti F, Kollmann K, et al. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene 2016;35:3083-91. [Crossref] [PubMed]
- Liu P, Begley M. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature 2014;508:541-5. [Crossref] [PubMed]
- Yeh HT, Tsai YS, Chen MS, et al. Flavopereirine induces cell cycle arrest and apoptosis via the AKT/p38 MAPK/ERK1/2 signaling pathway in human breast cancer cells. Eur J Pharmacol 2019;863:172658. [Crossref] [PubMed]
- Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet 2003;19:640-8. [Crossref] [PubMed]


