
Higher T cell immunoglobulin mucin-3 (Tim-3) expression in cervical cancer is associated with a satisfactory prognosis
Introduction
Cervical cancer (CC) had an estimated 570,000 new cases and 311,000 deaths globally in 2018, and it is the fourth most common gynecological cancer in the world, accounting for over 10% of all female cancers (1,2). A key cause of cervical intraepithelial neoplasia (CIN) and CC is high-risk human papillomavirus (HR-HPV) infection (3). More than 99% of CC are associated with HPV infection, primarily HPV-16 and HPV-18. There are three HPV vaccines that may be used to prevent CC: 2-valent, 4-valent, and 9-valent vaccines. However, the prognosis of patients diagnosed in the advanced stages still remains up for debate in regard to CC (4,5). The prognosis of CC patients is closely related to the patients’ FIGO stage, depth of cervical invasion and lymph node metastasis (6). Recently, RPA3, PD-1/PD-L1 and GINS2 were found to be the prognostic biomarkers of CC (7-10). In addition, molecular biomarkers put light on carcinogenic features that can be useful in developing novel anti-CC therapies (11,12). In this study, T cell immunoglobulin mucin-3 (Tim-3) is investigated as a prospective immune-checkpoint target, a topic still under debate.
Tim-3, a cell surface star molecule, consists of immunoglobulin and mucin-like domains and was first discovered in 2002 (13). A previous study reported that Tim-3 was expressed on a large number of immune cells like macrophages, dendritic cells, and T cells, where it was thought to be a novel immune regulator (14). Moreover, Tim-3 may be expressed on tumor cells (15). Another study demonstrated that the expression of Tim-3 is related to the prognosis of gastric cancer (16), hepatocellular carcinoma (17), renal cell carcinoma (18), breast cancer (19), and endometrial carcinoma (20). Recently, a particular study identified that Tim-3 expression in CC patients may promote metastatic potential (21), however, other studies reported that higher Tim-3 mRNA levels found in metastatic prostate cancer tissues served as an indicator of a favorable prognosis (22). Therefore, more comprehensive studies are needed to explore this dynamic.
The prognosis of CC patients in the advanced stages is still unsatisfactory. Targeted therapies confer clinical efficacy than cytokine therapies due to its direct inhibition of the targeted molecules. However, after a certain period of treatment, many patients become intractable to these therapies (23). Checkpoint inhibitors targeting cytotoxic T lymphocyte associated antigen 4 (CTLA-4) or programmed death receptor-1 (PD-1) have been introduced, and clinical trials are ongoing (24-26). To improve survival of patients with advanced CC, ideal therapy targets need to be explored.
In our study, first, we analyzed the medians of PD1, PDL1, CTLA4 and Tim-3 expression with the overall survival (OS) and found that only Tim-3 expression was obviously significant according to the median. We then investigated the relationship among the expression of Tim-3 and the clinicopathological variables. Next, we compared the OS according to Tim-3 expression and investigated the role of Tim-3 expression in tumors in the prognosis of CC based on the clinical parameters and RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) database. Finally, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene set enrichment analysis (GSEA) was performed on the TCGA data to analyze the biologically meaningful changes in CC and suggest possible treatment strategies in the comprehensive treatment of CC.
Methods
Patients and data
TCGA is a comprehensive publicly available collection of genomic data of more than 30 human tumors that provide researchers with data to improve the treatment of human cancers (27). The clinical characteristics, follow-up information, OS information and mRNA levels in tumors of patients with CC were abstracted from the TCGA dataset (http://cancergenome.nih.gov/). Accordingly, 294 samples of RNA-seq data were collected between 1994 and 2013 and were included in this study. The inclusion criteria were patients suffering from primary CC. Patients who were excluded from the study had incomplete information in regards to their staging or prognosis as well as any undetected genes.
GO annotation analysis
The GO project (http://www.geneontology.org) has been widely used to carry out the biologically meaningful annotation of two groups of Tim-3 mRNA levels.
KEGG pathway enrichment analysis
The enrichment analysis of KEGG pathway was done for the two groups of Tim-3 levels and was performed using the website (DAVID, http://david-d.ncifcrf.gov/).
GSEA
GSEA was carried out using the GSEA3.0 software (www.broad.mit.edu/gsea) to ascertain the pathways in the low Tim-3 expression group. All phenotype labels (.cls) and dataset (.gct) files were created and loaded into the GSEA software. RNA-seq data were downloaded from TCGA, which contained a total of 20,530 genes. During the GSEA analysis, genes were abstracted from the Molecular Signatures Database (MSigDB). MSigDB collects information on various kinds of genes, including 1,320 canonical pathways from KEGG, BioCarta, PID, Reactome, and other pathway databases (28). Data from TCGA were analyzed by GSEA, and pathways with an FDR <0.05 were considered significant. Herein, the permutations number was 1,000 and the phenotype label was Tim-3-low versus Tim-3-high.
Statistical analysis
All statistical analyses were performed using the SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Patients with CC were separated into two groups based on the median value of Tim-3 expression in tumor cells. Data from different groups were compared using the Student’s t-test and chi-square test or Fisher exact test. A Kaplan-Meier curve was conducted to examine whether the Tim-3 expression level in CC has prognostic value for the OS of patients with CC. Then, a univariate and multivariate logistic regression model was done to investigate whether Tim-3 expression was an independent prognostic factor in the OS. Moreover, a Spearman correlation analysis was performed to verify the relevance between immune-related genes and Tim-3 expression levels, and P<0.05 was regarded as statistically significant. Prism 7 (GraphPad Software Inc., La Jolla, USA) was used for all statistics related to clinical samples, while the R software (version 3.4; R Foundation for Statistical Computing, Vienna, Austria) was used for bioinformatics analyses.
Results
Description of the integrated CC data in TCGA
The integrated data of 287 CC patients from TCGA were enrolled for analysis. Of the 287 patients in the cohort, the median age of the CC patients was 46 years, ranging from 20 to 88 years. The clinical characteristics, follow-up information, tumor pathological features and OS information of the CC patients are presented in Table 1. The median follow-up time was 549 days among all CC patients, and 90 patients died during follow-up.
Table 1
| Characteristics | No. of cases | % |
|---|---|---|
| Age | ||
| ≤60 | 149 | 51.9 |
| >60 | 138 | 48.1 |
| BMI | ||
| <25 | 91 | 31.7 |
| ≥25 | 154 | 53.7 |
| Not recorded | 42 | 14.6 |
| Smoking history | ||
| Non-smoker | 139 | 48.4 |
| ≤15 years | 105 | 36.6 |
| >15 years | 9 | 3.1 |
| Not recorded | 34 | 11.9 |
| Total number of pregnancies | ||
| ≤5 | 213 | 74.2 |
| >5 | 41 | 14.3 |
| Not recorded | 33 | 11.5 |
| Number of successful birthed | ||
| ≤5 | 231 | 80.5 |
| >5 | 19 | 6.6 |
| Not recorded | 37 | 12.9 |
| Histological type | ||
| CSCC | 238 | 82.9 |
| Others | 49 | 17.1 |
| M | ||
| M0 | 105 | 36.6 |
| M1 | 10 | 3.5 |
| Mx | 125 | 43.6 |
| Not recorded | 47 | 16.3 |
| TNM stage | ||
| IA−IIA | 171 | 59.6 |
| IIB−IV | 105 | 36.6 |
| Stage X | 5 | 1.7 |
| Not recorded | 6 | 2.1 |
| Histologic grade | ||
| G1/2 | 147 | 51.2 |
| G3/4 | 114 | 39.7 |
| GX | 23 | 8.0 |
| Not recorded | 3 | 1.1 |
| Papillomavirus | ||
| HPV (−) | 25 | 8.7 |
| HPV (+) | 21 | 7.3 |
| Not recorded | 241 | 84.0 |
| Hysterectomy type | ||
| Simple/radical hysterectomy | 161 | 56.1 |
| Other | 126 | 43.9 |
| Survival status | ||
| Alive | 217 | 75.6 |
| Died | 70 | 24.4 |
CSCC, cervical squamous cell carcinoma; BMI, body mass index; G, histologic grade.
The Tim-3 mRNA levels of the CC patients were downloaded from the RNA-seq data, where the median of Tim-3 in CC patients was found to be 7.7168 ng/mL, containing continuous variables in a wide range of 3.823 to 11.1941 ng/mL.
K-M survival curves of Tim-3
The CC patients were further divided into a high Tim-3 group and a low Tim-3 group according to the median Tim-3 mRNA value. A low expression of Tim-3 in tumor tissues was associated with a poor prognosis (P=0.0431), as shown in Figure 1A. The 5-year OS rate of patients with early CC was obviously longer than those with advanced CC (P<0.0001), as depicted in Figure 1B. Similarly, the 5-year OS rate of patients with a lower tumor size (T), node (N) and metastasis (M) was obviously longer than those with higher T, N and M stage (P<0.0001, P=0.0044 and P=0.0052), illustrated in Figures 1C,D,E. The corresponding results imply that Tim-3 may serve as a satisfactory prognostic biomarker in CC.
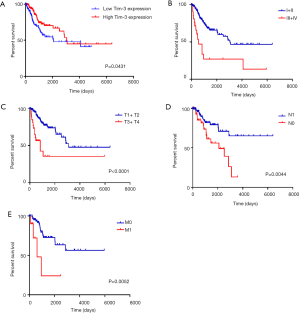
Clinical features of the integrated CC patients
We investigated the relationship between the expression of Tim-3 and the clinicopathological variables. The results demonstrated a correlation between high expression levels of Tim-3 in patients with CC smoking, BMI, FIGO stage, metastasis and histological type, as seen in Figures 2A,B,C,D,E,F. Moreover, we found that the high expression of Tim-3 was strongly association with the smoking history (P=0.012), total number of pregnancies (P=0.002), histological type (P<0.0001), M stage (P=0.036), FIGO stage (P=0.001), papillomavirus (P=0.001), hysterectomy type (P<0.0001) and survival status (P<0.0001), depicted in Table 2. Therefore, we identified Tim-3 as a prognostic gene associated with tumor progression in CC patients. Furthermore, logistic regression analysis was done to examine Tim-3’s usefulness in the prognosis of CC. Univariate analyses revealed that the T stage (HR =3.80; P=0.001), N stage (HR =2.59; P=0.001), M stage (HR =4.12; P=0.001), TNM stage (HR =1.86; P=0.001) and high Tim-3 levels (HR =1.62; P=0.04) were associated with the survival of CC patients. Additionally, multivariate analyses showed that the histological type (HR =0.13; P=0.02), TNM stage (HR =3.01; P=0.04) and high Tim-3 levels (HR =3.41; P=0.04) were independent predictors of prognosis, which are detailed in Table 3. Collectively, our results suggest that the expression of Tim-3 plays a critical role in the prognosis of CC patients.
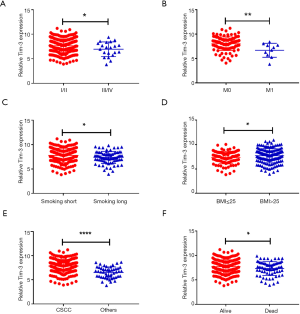
Table 2
| Characteristics | Total, N (%) | Tim-3 expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Age | 0.955 | |||
| ≤46 | 149 | 74 | 75 | |
| >46 | 138 | 69 | 69 | |
| BMI | 0.082 | |||
| <25 | 91 | 38 | 53 | |
| ≥25 | 154 | 82 | 72 | |
| Not recorded | 42 | 23 | 19 | |
| Smoking history | 0.012* | |||
| Non-smoker | 139 | 82 | 57 | |
| ≤15 years | 105 | 45 | 60 | |
| >15 years | 9 | 4 | 5 | |
| Not recorded | 34 | 12 | 22 | |
| Total number of pregnancies | 0.002** | |||
| ≤5 | 213 | 104 | 109 | |
| >5 | 41 | 25 | 16 | |
| Not recorded | 33 | 13 | 20 | |
| Number of successful birthed | 0.756 | |||
| ≤5 | 231 | 118 | 113 | |
| >5 | 19 | 9 | 10 | |
| Not recorded | 37 | 15 | 22 | |
| Histological type | <0.0001**** | |||
| CSCC | 238 | 136 | 102 | |
| Others | 49 | 7 | 42 | |
| M | 0.036*,b | |||
| M0 | 105 | 67 | 38 | |
| M1 | 10 | 3 | 7 | |
| Mx | 125 | 59 | 66 | |
| Not recorded | 47 | 14 | 33 | |
| FIGO stage | 0.001* | |||
| IA-IIA | 171 | 101 | 70 | |
| IIB-IV | 105 | 40 | 65 | |
| Stage X | 5 | 0 | 5 | |
| Not recorded | 6 | 2 | 4 | |
| Histologic grade | 0.067 | |||
| G1/2 | 147 | 67 | 80 | |
| G3/4 | 114 | 65 | 49 | |
| GX | 23 | 9 | 14 | |
| Not recorded | 3 | 2 | 1 | |
| Papillomavirus | 0.001** | |||
| HPV (−) | 25 | 12 | 13 | |
| HPV (+) | 41 | 35 | 6 | |
| Not recorded | 221 | 96 | 125 | |
| Hysterectomy type | <0.0001**** | |||
| Simple/radical hysterectomy | 161 | 90 | 71 | |
| Other | 126 | 53 | 73 | |
| Survival status | <0.0001**** | |||
| Alive | 197 | 114 | 83 | |
| Died | 90 | 29 | 61 | |
b, indicates the Fisher test, and the other test was the Chi-square test. *, P<0.05; **, P<0.01; ***, P<0.001****, P<0.0001. CSCC, cervical squamous cell carcinoma; BMI, body mass index; G, histologic grade.
Table 3
| Characteristics | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (>46 vs. ≤46) | 1.35 | 0.59–3.17 | 0.41 | 1.39 | 0.87–3.45 | 0.67 | |
| BMI (<25 vs. ≥25) | 1.72 | 0.97–3.04 | 0.05 | 0.48 | 0.14–1.61 | 0.23 | |
| Smoking years (>15 to ≤15) | 0.95 | 0.31–2.95 | 0.93 | 1.30 | 0.01–119.00 | 0.91 | |
| Total number of pregnancies (≥5 vs. <5) | 1.23 | 0.63–2.41 | 0.51 | 2.95 | 0.52–16.71 | 0.22 | |
| Number of successful birthed (≥5 vs. <5) | 1.24 | 0.49–3.11 | 0.62 | 0.02 | 0.01–0.03 | 0.99 | |
| Histological type (CSCC vs. others) | 1.03 | 0.55–1.96 | 0.91 | 0.13 | 0.02–0.74 | 0.02* | |
| T (T3/4 vs. T1/2) | 3.80 | 1.47–9.86 | 0.001*** | 0.13 | 0.04–4.04 | 0.25 | |
| N (N1 vs. N0) | 2.59 | 1.22–5.49 | 0.001** | 1.49 | 0.30–7.34 | 0.63 | |
| M (M1 vs. M0) | 4.12 | 1.712–8.38 | 0.001** | 3.44 | 1.68–6.13 | 0.001** | |
| TNM stage (IIB-IV vs. IA-IIA) | 1.86 | 1.13–3.08 | 0.001** | 3.01 | 1.03–8.82 | 0.04* | |
| G (G1/2 vs. G3/4) | 1.16 | 0.70–1.95 | 0.57 | 0.73 | 0.23–2.37 | 0.60 | |
| Papillomavirus (+ vs. −) | 1.11 | 0.33–3.72 | 0.87 | 0.89 | 0.16–3.28 | 0.07 | |
| Hysterectomy type (simple/radical hysterectomy vs. others) | 0.70 | 0.13–3.76 | 0.62 | 0.89 | 0.45–3.32 | 0.86 | |
| Radiotherapy (yes vs. no) | 1.06 | 0.60–1.87 | 0.85 | 0.63 | 0.13–5.16 | 0.63 | |
| Chemotherapy (yes vs. no) | 0.94 | 0.12–7.27 | 0.95 | 3.12 | 0.39–4.26 | 0.76 | |
| Targeted molecular therapy (yes vs. no) | 0.94 | 0.54–1.65 | 0.83 | 2.22 | 0.68–3.21 | 0.65 | |
| Tim-3 (low vs. high) | 1.62 | 1.43–3.67 | 0.04* | 3.41 | 1.56–12.33 | 0.04* | |
CSCC, cervical squamous cell carcinoma; BMI, body mass index; G, histologic grade. *, P<0.05; **, P<0.01; ***, P<0.001.
Significant GOs and pathways
To predict the biological function of Tim-3 in the progression of CC, a GO enrichment analysis and a KEGG analysis were performed. The results demonstrated Tim-3’s involvement in various biological processes. Figure 3A,B depict the associated functional pathways, where it is observed that Tim-3 closely relates to macrophage differentiation, monocyte chemotaxis, macrophage activation, lymphocyte mediated immunity, leukocyte differentiation and interleukin 2 production. In order to identify the key pathways, a pathway analysis was performed, where it was found that the major regulated biological pathways are B cell receptor signaling, ECM receptor interaction, T cell receptor signaling, cell cycle, apoptosis, cell adhesion molecules and JAK-STAT signaling, implying that Tim-3 potentially regulates the immune system.
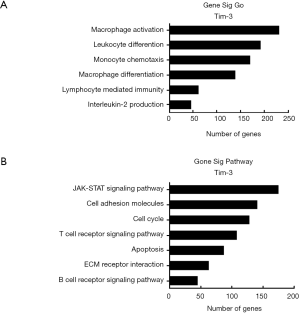
GSEA analysis of Tim-3
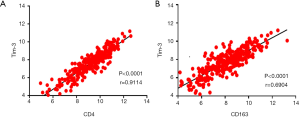
Based on the above results, we discovered that Tim-3 plays a crucial role in the prognosis of CC. Consequently, it was necessary to explore the biological functions of Tim-3. GSEA, a powerful tool used to estimate biological functions, was utilized in this regard. The results showed that Tim-3 expression was associated with macrophage differentiation, regulation of monocyte chemotaxis, negative regulation of interleukin 2 production, positive regulation of substrate adhesion dependent cell spreading, regulation of NF kappa-B signaling, STAT cascade, erk1 and erk2 cascade and regulation of vascular endothelial growth factor receptor signaling pathway. These functions were significantly enriched at low levels of Tim-3 expression in CC, as seen in Figure 4. This demonstrated that Tim-3 expression levels may serve as a prognostic indicator in CC patients with favorable outcomes. Then, a database search was done for the expression profiles of Tim-3 as well as any immune-related genes, where a positive correlation was observed between Tim-3 expression and the expression of immune markers, such as CD4 (P<0.0001, r=0.9114) and CD163 (P<0.0001, r=0.6904), as illustrated in Figures 5A,B. Seemingly, a high level of Tim-3 confers an immunological tumor-promoting role. Essentially, our study infers that a high level of Tim-3 is closely associated with a favorable prognosis in CC patients in regard to their immunity.
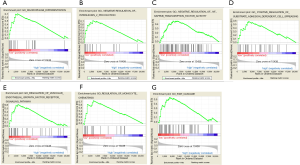
Discussion
Thanks to the increasing availability of high-throughput technologies, many novel therapeutic targets and biomarkers have been identified through the transcriptomic analysis of various tumors. In this paper, our results imply that high Tim-3 mRNA levels in CC are associated with a satisfactory prognosis via activation of immune responses in cervical tumor tissues. The identification of CC biomarkers may help predict and prolong the survival of CC patients and may serve as an ideal therapeutic target.
Tim-3 is known to be a novel immune regulator that is expressed in a variety of tumor tissues and is closely related with tumor development (17,29). High levels of Tim-3 expression in peripheral NK cells (30), tumor infiltrating T cells (31,32), macrophages/monocytes (33), contributed to an unfavorable prognosis in a variety of cancers. However, in this study, we found that downregulation of Tim-3 indicates a worse outcome. The underlying mechanism explaining the according results lies in that the Tim-3 expressed in tumor cells and tumor-associated immune cells exhibit different tumorigenic patterns (34). Therefore, conflicting results pertaining to the relationship between Tim-3 expression in tumor cells and the survival data in different cancers were obtained (21,22). The upregulation of Tim-3 in esophageal cancer indicated a shorter survival of patients (35), while a higher expression of Tim-3 may be a protective factor in patients with colon cancer (36). This complicated phenomenon may demonstrate that the localization of varying levels of Tim-3 expression encompass different functions in different tumor tissues.
Our study provides new insight into the prognostic value of Tim-3 in patients with CC. Accordingly, we obtained CC data from TCGA and evaluated the different expression levels of Tim-3 in CC tissues as well as the clinicopathological variables. We found that Tim-3 is a favorable prognostic biomarker for patients with CC, and a logistic regression analysis suggests that the expression level of Tim-3 plays an important role in predicting the prognosis of CC patients. According to the pathway analysis, the underlying mechanism may deal with macrophage differentiation, macrophage activation, lymphocyte mediated immunity, regulation of monocyte chemotaxis, negative regulation of interleukin 2 production, positive regulation of substrate adhesion dependent cell spread and other mechanisms.
However, the results obtained in this study contrasts with a previous study suggesting that Tim-3 expression in patients with CC may be attributed with increased metastatic potential (21). Hence, controversial results in regard to the association between Tim-3 expression levels in CC cells and CC patient prognosis were obtained. This phenomenon suggests that the role of Tim-3 in CC remains ambiguous. Moreover, the underlying reason for these results has not been fully clarified. Therefore, further investigation and experimental validation are needed in order to elucidate the exact role of Tim-3 in patients with CC in order to develop a more accurate treatment plan.
In summary, this report first indicated that Tim-3 expression was downregulated in CC tissues in advanced stages compared to those in early stages. The absence of Tim-3 contributed to tumor progression, resulting in poor patient outcomes. This is the first study showing that Tim-3 exerts a detrimental effect on the progression of CC via bioinformatic screening. The expression levels of Tim-3 may be a valuable adjuvant parameter to predict the prognosis of patients with CC and provides a potential therapeutic foundation.
This study has the following limitations: no clinical sample was verified, and different types of CC were included. Therefore, further investigations should focus on the observation of the differences between expression levels and prognosis in CC.
Conclusions
To conclude, a higher level of expression of Tim-3 serves as a biomarker in the favorable prognosis of CC. Hence, Tim-3 has the potential to shape the landscape of immunotherapy for CC.
Acknowledgments
Funding: This paper was supported by grants from
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure from (available at http://dx.doi.org/10.21037/tcr.2020.02.41). The authors have no conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dai F, Chen G, Wang Y, et al. Identification of candidate biomarkers correlated with the diagnosis and prognosis of cervical cancer via integrated bioinformatics analysis. Onco Targets Ther 2019;12:4517-32. [Crossref] [PubMed]
- Celewicz A, Celewicz M, Wezowska M, et al. Clinical efficacy of p16/Ki-67 dual-stained cervical cytology in secondary prevention of cervical cancer. Pol J Pathol 2018;69:42-7. [Crossref] [PubMed]
- Park E, Kim JY, Choi S, et al. Carcinogenic risk of human papillomavirus (HPV) genotypes and potential effects of HPV vaccines in Korea. Sci Rep 2019;9:12556. [Crossref] [PubMed]
- Xie F, Dong D, Du N, et al. An 8gene signature predicts the prognosis of cervical cancer following radiotherapy. Mol Med Rep 2019;20:2990-3002. [PubMed]
- Chen S, Tao M, Zhao L, et al. The association between diabetes/hyperglycemia and the prognosis of cervical cancer patients: A systematic review and meta-analysis. Medicine (Baltimore) 2017;96:e7981. [Crossref] [PubMed]
- Sun L, Jiang R, Li J, et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem 2017;54:127-33. [Crossref] [PubMed]
- Maleki Vareki S, Garrigos C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol 2017;116:116-24. [Crossref] [PubMed]
- Ouyang F, Liu J, Xia M, et al. GINS2 is a novel prognostic biomarker and promotes tumor progression in early-stage cervical cancer. Oncology Reports 2017;37:2652-62. [Crossref] [PubMed]
- Qu C, Zhao Y, Feng G, et al. RPA3 is a potential marker of prognosis and radioresistance for nasopharyngeal carcinoma. J Cell Mol Med 2017;21:2872-83. [Crossref] [PubMed]
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22:813-20. [Crossref] [PubMed]
- Kambayashi Y, Fujimura T, Hidaka T, et al. Biomarkers for Predicting Efficacies of Anti-PD1 Antibodies. Front Med (Lausanne) 2019;6:174. [Crossref] [PubMed]
- de Mingo Pulido A, Gardner A, Hiebler S, et al. TIM-3 Regulates CD103(+) Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018;33:60-74.e6. [Crossref] [PubMed]
- Yan W, Liu X, Ma H, et al. Tim-3 fosters HCC development by enhancing TGF-beta-mediated alternative activation of macrophages. Gut 2015;64:1593-604. [Crossref] [PubMed]
- Friedlaender A, Addeo A, Banna G. New emerging targets in cancer immunotherapy: the role of TIM3. ESMO Open 2019;4:e000497. [Crossref] [PubMed]
- Yu J, Zhang H, Sun S, et al. The effects of Tim-3 activation on T-cells in gastric cancer progression. Oncol Lett 2019;17:1461-6. [PubMed]
- Liu F, Liu Y, Chen Z. Tim-3 expression and its role in hepatocellular carcinoma. J Hematol Oncol 2018;11:126. [Crossref] [PubMed]
- Zhang X, Yin X, Zhang H, et al. Differential expression of TIM-3 between primary and metastatic sites in renal cell carcinoma. BMC Cancer 2019;19:49. [Crossref] [PubMed]
- Burugu S, Gao D, Leung S, et al. TIM-3 expression in breast cancer. Oncoimmunology 2018;7:e1502128. [Crossref] [PubMed]
- Moore M, Ring KL, Mills AM. TIM-3 in endometrial carcinomas: an immunotherapeutic target expressed by mismatch repair-deficient and intact cancers. Mod Pathol 2019;32:1168-79. [Crossref] [PubMed]
- Cao Y, Zhou X, Huang X, et al. Tim-3 expression in cervical cancer promotes tumor metastasis. PLoS One 2013;8:e53834. [Crossref] [PubMed]
- Wu J, Lin G, Zhu Y, et al. Low TIM3 expression indicates poor prognosis of metastatic prostate cancer and acts as an independent predictor of castration resistant status. Sci Rep 2017;7:8869. [Crossref] [PubMed]
- Nishijima TF, Shachar SS, Nyrop KA, et al. Safety and Tolerability of PD-1/PD-L1 Inhibitors Compared with Chemotherapy in Patients with Advanced Cancer: A Meta-Analysis. Oncologist 2017;22:470-9. [Crossref] [PubMed]
- De Felice F, Marchetti C, Palaia I, et al. Immune check-point in cervical cancer. Crit Rev Oncol Hematol 2018;129:40-3. [Crossref] [PubMed]
- Tamura K, Hasegawa K, Katsumata N, et al. Efficacy and safety of nivolumab in Japanese patients with uterine cervical cancer, uterine corpus cancer, or soft tissue sarcoma: Multicenter, open‐label phase 2 trial. Cancer Science 2019;110:2894-904. [Crossref] [PubMed]
- Rotman J, Mom CH, Jordanova ES, et al. 'DURVIT': a phase-I trial of single low-dose durvalumab (Medi4736) IntraTumourally injected in cervical cancer: safety, toxicity and effect on the primary tumour- and lymph node microenvironment. BMC Cancer 2018;18:888. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Liu J, Wang D, Zhang C, et al. Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling. Chin J Cancer Res 2018;30:633-46. [Crossref] [PubMed]
- Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunological Reviews 2017;276:97-111. [Crossref] [PubMed]
- Xu L, Huang Y, Tan L, et al. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol 2015;29:635-41. [Crossref] [PubMed]
- Cai C, Xu YF, Wu ZJ, et al. Tim-3 expression represents dysfunctional tumor infiltrating T cells in renal cell carcinoma. World J Urol 2016;34:561-7. [Crossref] [PubMed]
- Zhang X, Zhang H, Chen L, et al. TIGIT expression is upregulated in T cells and causes T cell dysfunction independent of PD-1 and Tim-3 in adult B lineage acute lymphoblastic leukemia. Cell Immunol 2019;344:103958. [Crossref] [PubMed]
- Wang Z, Yin N, Zhang Z, et al. Upregulation of T-cell Immunoglobulin and Mucin-Domain Containing-3 (Tim-3) in Monocytes/Macrophages Associates with Gastric Cancer Progression. Immunol Invest 2017;46:134-48. [Crossref] [PubMed]
- Kang CW, Dutta A, Chang LY, et al. Apoptosis of tumor infiltrating effector TIM-3+CD8+ T cells in colon cancer. Sci Rep 2015;5:15659. [Crossref] [PubMed]
- Shan B, Man H, Liu J, et al. TIM-3 promotes the metastasis of esophageal squamous cell carcinoma by targeting epithelial-mesenchymal transition via the Akt/GSK-3beta/Snail signaling pathway. Oncol Rep 2016;36:1551-61. [Crossref] [PubMed]
- Sun QY, Qu CH, Liu JQ, et al. Down-regulated expression of Tim-3 promotes invasion and metastasis of colorectal cancer cells. Neoplasma 2017;64:101-7. [Crossref] [PubMed]


