
Interaction between linc01615 and miR-491-5p regulates the survival and metastasis of colorectal cancer cells
Introduction
Colorectal cancer (CRC) is the third most common cancer and fourth leading cause of cancer death worldwide. The incidence of CRC has been rapidly rising in many Asian countries in the past few decades (1). Approximately 21% of CRC are diagnosed with the existence of metastasis, and these patients have a 5-year survival rate of 13.8% in the United States in 2018 (2). However, there is a limited improvement in the 5-year survival during the past several decades (2). During tumorigenesis, the normal epithelium of the gastrointestinal tract becomes hyperproliferative mucosa, and then becomes carcinoma which is associated with sequential activation of specific oncogenes and/or inactivation of tumor suppressor genes (3). However, CRCs are very heterogeneous in both the biological behavior and molecular changes. Thus, exploring new molecular markers is still significant for understanding the tumorigenesis of CRCs.
Long intergenic non-coding RNAs (lincRNAs) are autonomously transcribed non-coding RNAs that are longer than 200 nucleotides (4). LincRNAs function in regulating chromatin topology and neighboring genes, scaffolding proteins and other RNAs, acting as protein and RNA decoys, and subsequently being involved in tumorigenesis (5). LincRNAs have high tissue specificity and, thus, are proposed to fine-tune the expression of their target genes in a tissue-specific manner (6). For example, linc00152 has been demonstrated to contribute to the carcinogenesis of several cancers in the digestive system through regulating various signaling pathways, and high linc00152 expression is a biomarker for chemoresistance and poor prognosis of patients with these cancers (7). A recent study proposed that linc01615 may potentially affect the extracellular matrix and subsequently have an impact on the metastasis of hepatocellular carcinoma (8). However, whether or not linc01615 is involved in the survival and metastasis of CRC has not been reported, and especially, how the linc01615 was regulated and what genes it regulates still remains to be identified.
MicroRNAs (miRNAs) are short non-coding RNAs that are involved in posttranscriptional regulation of gene expression. miRNAs have been demonstrated to be important in all biological processes, and aberrant miRNA expression is involved in many diseases including cancer (9). The regulative effects of miRNAs on lincRNA have recently been proposed to promote the degeneration of lincRNAs (10). In contrast, several lincRNAs have been reported to regulate miRNAs expression in colorectal cancer. For instance, linc00152 modulates the expression of miR-193a-3p (11); linc01567 negatively regulates the expression of miRNA-93 in colon cancer stem cells (12); linc01410 inhibits miR-3128 expression in colon cancer cells (13); and knockdown of lincRNA-ROR restores the expression of miR-145 (14).
In this study, we investigated the regulative effects of 9 putative miRNAs on the expression of linc01615. In contrast, we also tested whether linc01615 also regulates the expression of the selective miRNA in colorectal cancer cells.
Methods
Cell culture
Six human gastric colorectal cancer cell lines (SW480, SW620, HCT-116, HT29, CaCo-2, and LoVo1) and one normal human colon mucosal epithelial cell line (NCM460) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 medium supplied with 10% FBS (fetal bovine serum) (Thermo Fisher Scientific, Waltham, MA USA) at 37 °C, 5% CO2.
Real-time PCR
Cellular total RNA was isolated from cultured cells and purified using miRNeasy Mini Kit (Qiagen #217004, Germantown, MD, USA). microRNA was amplified in the Bio-Rad CFX96™ Real-time system (Bio-Rad Laborateries, Hercules, CA, USA) using miRNA Q-PCR Detection kit (GeneCopoeia #R0101L, Rockville, MD, USA). The primers were listed in Table 1. To amplify the linc01615 gene, total RNA was extracted using Trizol reagent (Fermentas #K1631, Waltham, MA, USA), and PCR was performed using SYBR Green PCR Master Mix (ThermoFisher Scientific #4309155, Waltham, MA USA). Linc01615 and β-actin gene (internal control) were amplified using forward and reverse primer (Table 1). Data were analyzed using 2−ΔΔCt (RQ: Relative quantification).
Table 1
| miRNA | Primer | Sequence 5'-3' |
|---|---|---|
| hsa-miR-590-3p | Forward | ACACTCCAGCTGGGTAATTTTATGTATAA |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTAGCTT | |
| hsa-miR-455-3p | Forward | ACACTCCAGCTGGGGCAGTCCATGGGCAT |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTGTATAT | |
| hsa-miR-491-5p | Forward | ACACTCCAGCTGGGAGTGGGGAACCCTTCC |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTCATGG | |
| hsa-miR-484 | Forward | ACACTCCAGCTGGGTCAGGCTCAGTCCCCT |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATCGGGAG | |
| hsa-miR-339-5p | Forward | ACACTCCAGCTGGGTCCCTGTCCTCCAGGAG |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGTGAGCT | |
| hsa-miR-1296-5p | Forward | ACACTCCAGCTGGGTTAGGGCCCTGGCTCC |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGAGATGG | |
| hsa-miR-22-3p | Forward | ACACTCCAGCTGGGAAGCTGCCAGTTGAAG |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGTTCT | |
| hsa-mir-4316 | Forward | ACACTCCAGCTGGGGGTGAGGCTAG |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCACCAGCT | |
| hsa-miR-1301-3p | Forward | ACACTCCAGCTGGGTTGCAGCTGCCTGGGAGT |
| Reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAAGTCAC | |
| U6snRNA | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
Linc01615 siRNA design and overexpression vector construction
Three siRNA sequences were designed to target linc01615 gene (siLINC1, siLINC2, siLINC3) (Table 1). A non-targeting small RNA was used as a control (Table 1). To construct the overexpression vector, the human linc01615 gene was amplified using forward primer: 5'-CTGGCCGTGGGGTCAGCTGCAC-3' and reverse primer: 5'-ACGCAGGTTTTGTTGCGAAGTG-3' and directly cloned into pcDNA™3.1/V5-His™ TOPO™ TA Expression vector (ThermoFisher Scientific #K480001, Waltham, MA USA). The sequence was confirmed by sequencing. The miR-491-5p mimic was synthesized according to the sequence of matured miR-491-5p (miR-491 m), whereas miR-491-5p inhibitor (miR-491 In) was an antisense sequence of miR-491-5p (Ruibo Biological Technology Co., Ltd, Guangzhou, China).
MTT assay
A total of 4×103 Caco-2 cells suspended in 200 µL of medium were seeded into each well of 96-well plates and cultured overnight. Cells were then transfected with linc01615 siRNA (siLINC), linc01615 overexpression vector (oeLINC), miR-491-5p mimics (miR-491 m), and miR-491-5p inhibitor (miR-491 In). After culturing for 24 h, 20 µL/well MTT [3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), 5 mg/mL] (Sigma-Aldrich, M5655, St. Louis, MO, USA) was added and incubated for 4 h. Two hundred µL/well of isopropanol was then added, and the plates were shaken for 10 min at room temperature. The absorbance was read at 490 nm.
Invasion assay
Transwell chamber (Corning Life Science #3428, New York, USA) was covered with a membrane (8 µM holes) and coated with 100-µL of Matrigel Matrix (Corning Life Science #354248) mixed with culture medium (1:8 dilution). One chamber was then placed in a 6 cm dish. Caco-2 cells were then transfected with shLINC, oeLINC, miR-491 m, miR-491 In, or controls for 24 h. After trypsinized. A total of 1.0×105 cells were resuspended in 500 µL of serum-free medium and seeded in the chambers, while 1 mL of medium containing 10% FBS was added into the well. After a continuous culture of 24 h, the membranes were fixed with 95% alcohol and stained with 2% crystal violet solution. After washing with 1× PBS, cells on the membranes were observed under an inverted microscope.
Cell scratch test
Caco-2 cells were treated as described above and resuspended in DMEM medium containing 5% FBS. 5.0×105 cells were plated onto the 6-cm dish to create a confluent monolayer. When the cell fluency was 100%, a scratch was made in a straight line with a pipet tip. After removing the debris, cells were continuously cultured at 37 °C for 48 h. The dishes were examined periodically and observed under a phase-contrast microscope, and the images were analyzed using Image Pro-Plus software (Media Cybernetics, Sliver Spring, MD, USA).
Statistical analysis
Data were analyzed using SPSS 15.0 statistical software (IBM, USA). One-way ANOVA was used to compare differences between groups. A P<0.05 was considered statistically significant.
Results
The mRNA expression of linc01615 gene in colorectal cancer cells
The mRNA expression of human linc01615 gene in six colorectal cancer cell lines and one normal colon mucosal epithelial cell line was analyzed using real-time PCR. Among the six cancer cell lines, the highest linc01615 mRNA (Figure 1A) expression was observed in Caco-2 cells. After treating the Caco-2 cells with three siRNAs of linc01615 gene (siLINC1, siLINC2, siLINC3), real-time PCR showed that siLINC2 was most effective in inhibiting LINC01615 expression, whereas the overexpression vector (oeLINC) significantly increased linc01615 mRNA expression (Figure 1B).
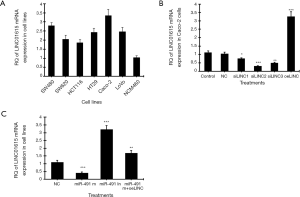
We further tested the effect of linc01615 on the expression of nine microRNAs in Caco-2 cells. These nine microRNAs were selected through sequence analysis and publications. Real-time PCR showed that silencing of linc01615 expression was the most effective in increasing miR-491-5p expression, whereas overexpressing of linc01615 was the most effective in decreasing miR-491-5p expression compared to other microRNAs (Table 2). In contrast, we also tested the effect of miR-491-5p on linc01615 mRNA expression in Caco-2 cells. Transfection with miR-491-5p mimics (miR-491 m) significantly downregulated linc01615 mRNA expression, whereas transfection with miR-491-5p inhibitor significantly increased linc01615 expression, and transfection with the combined miR-491-5p mimics and linc01615 overexpression vector (miR-491 m+oeLINC) significantly blocked the inhibitory effect of miR-491 m on linc01615 mRNA expression (Figure 1C).
Table 2
| Treatments | miR-590-3p | miR-455-3p | miR-491-5p | miR-484 | miR-339-5p | miR-1296-5p | miR-22-3p | miR-4316 | miR-1301-3p |
|---|---|---|---|---|---|---|---|---|---|
| Control | 1.1055 | 1.0809 | 1.0107 | 1.1422 | 1.1060 | 1.0446 | 1.0850 | 1.0807 | 1.1787 |
| NC | 1.0140 | 1.1594 | 1.0844 | 1.0423 | 1.1221 | 1.1200 | 0.9779 | 1.0449 | 1.0602 |
| siLINC01615 | 1.0934 | 2.3077 | 3.7138 | 2.5374 | 1.6499 | 1.3043 | 1.5641 | 0.9639 | 1.1409 |
| oeLINC01615 | 1.0883 | 0.6013 | 0.3903 | 0.6287 | 0.7955 | 0.8621 | 0.7102 | 1.0370 | 1.1230 |
LINC01615 and miR-491-5p expression affects the proliferation, apoptosis, invasion, and migration in Caco-2 cells
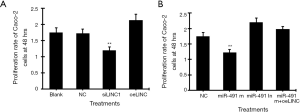
MTT assay showed that transfection of linc01615 siRNA (siSLINC) significantly decreased, whereas transfection of linc01615 overexpression vector (oeLINC) significantly increased cell proliferation in Caco-2 cells (Figure 2A). Similarly, transfection of miR-491-5p mimic (miR-491 m) significantly decreased, whereas transfection of miR-491-5p inhibitor (miR-491 In) significantly increased, cell proliferation in Caco-2 cells (Figure 2B). Co-transfection of miR-491 m with oeLINC significantly blocked the inhibitory effect of miR-491 m (Figure 2B).
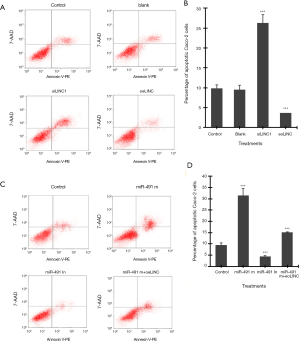
Flow cytometry assay showed that transfection of siLINC significantly increased, whereas transfection of oeLINC significantly decreased, cell apoptosis in Caco-2 cells (Figure 3A,B). Similarly, transfection of miR-491 m significantly increased, whereas transfection of miR-491 In significantly decreased, cell apoptosis in Caco-2 cells (Figure 3C,D). Co-transfection of miR-491 m with oeLINC significantly blocked the apoptotic effect of miR-491 m (Figure 3).
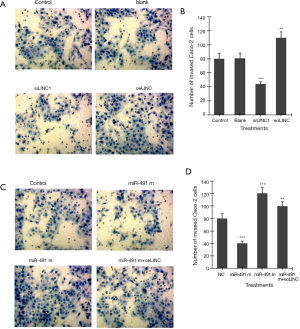
Transwell invasion assay showed that transfection of siLINC significantly decreased, whereas transfection of oeLINC significantly increased, invasion in Caco-2 cells (Figure 4A,B). Transfection of miR-491 m significantly decreased, whereas transfection of miR-491 In significantly increased, invasion in Caco-2 cells (Figure 4C,D). Co-transfection of miR-491 m with oeLINC significantly blocked the effect of miR-491 m on cell invasion (Figure 4).
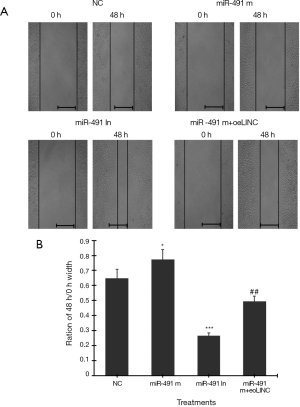
Cell scratch test showed that transfection of miR-491 m for 48 h significantly increased, whereas transfection of miR-491 In significantly decreased, the ratio of 48-h/0-h scratch width in Caco-2 cells (Figure 5A,B). Co-transfection of miR-491 m with oeLINC significantly blocked the effect of miR-491 m on cell migration (Figure 5).
Discussion
Although the therapy of CRC has been developed, the 5-year survival rate still has had not being significantly improved during the past several decades. This suggests that we are still far from uncovering the molecular mechanisms driving the progression of CRC. LincRNAs and/or lncRNAs are non-coding RNAs with regulative effects on genes’ expression. The functions of linc01615 and its regulation in CRC is rarely reported. In this study, we found that linc01615 affects the survival and metastasis of Caco-2 colorectal cancer cells through regulating miR-491-5p expression. In contrast, miR-491-5p also feedback regulated linc01615 expression in Caco-2 cells.
This study demonstrated that linc01615 mRNA expression was upregulated in six colorectal cancer cell lines compared to the normal colon mucosal epithelial cell line. Among the six tested CRC cell lines, Caco-2 cells exhibited the highest linc01615 expression, the miR-491-5p mimic significantly decreased linc01615 expression, and miR-491-5p inhibitor significantly increased linc01615 expression, suggesting that miR-491-5p negatively regulated linc01615 expression. In contrast, silencing of linc01615 expression significantly increased miR-491-5p expression while overexpression of linc01615 significantly decreased miR-491-5p expression in Caco-2 colorectal cancer cells; this suggests that linc01615 negatively regulated miR-491-5p expression. Thus, we can conclude that linc01615 and miR-491-5p can negatively regulate each other.
The biological activity analysis showed that silencing of linc01615 expression significantly decreased, whereas overexpression of linc01615 gene significantly increased, cell proliferation in colorectal cancer cells. In contrast, silencing of linc01615 expression significantly increased, whereas overexpression of linc01615 gene significantly decreased, cell apoptosis in colorectal cancer cells (Figures 1,2). These findings suggest that linc01615 functions as an oncogene in colorectal cancer cells. Conversely, transfection with miR-491-5p mimic significantly decreased cell proliferation and increased apoptosis in Caco-2 cells, suggesting that miR-491-5p is a tumor suppressor. Furthermore, the linc01615 and miR-491-5p play an opposite role in the invasion and migration of Caco-2 cells. The biological activities of miR-491-5p can be blocked by overexpression of linc01615 expression. In addition to the regulative effect of linc01615 on miR-491-5p expression, we concluded that linc01615 and miR-491-5p are a pair of genes to regulate each other tightly in colorectal cancer cells.
In conclusion, linc01615 functions as an oncogene, whereas miR-491-5p functions as a tumor suppressor, in colorectal cancer cells; they do so through negatively regulating each other. Both the linc01615 and miR-491-5p are involved in cell proliferation, apoptosis, invasion, and migration of colorectal cancer cells.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Veettil SK, Lim KG, Chaiyakunapruk N, et al. Colorectal cancer in Malaysia: Its burden and implications for a multiethnic country. Asian J Surg 2017;40:481-9. [Crossref] [PubMed]
- Patel JN, Fong MK, Jagosky M. Colorectal Cancer Biomarkers in the Era of Personalized Medicine. J Pers Med 2019;9:E3. [Crossref] [PubMed]
- De Rosa M, Pace U, Rega D, et al. Genetics, diagnosis and management of colorectal cancer Oncol Rep 2015;34:1087-96. (Review). [Crossref] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19:143-57. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Zhang Y, Tang L. The Application of lncRNAs in Cancer Treatment and Diagnosis. Recent Pat Anticancer Drug Discov 2018;13:292-301. [Crossref] [PubMed]
- Xu J, Guo J, Jiang Y, et al. Improved characterization of the relationship between long intergenic non-coding RNA Linc00152 and the occurrence and development of malignancies. Cancer Med 2019;8:4722-31. [Crossref] [PubMed]
- Ji D, Chen GF, Liu X, et al. Identification of LINC01615 as potential metastasis-related long noncoding RNA in hepatocellular carcinoma. J Cell Physiol 2019;234:12964-70. [Crossref] [PubMed]
- Acunzo M, Romano G, Wernicke D, et al. MicroRNA and cancer--a brief overview. Adv Biol Regul 2015;57:1-9. [Crossref] [PubMed]
- Tang XJ, Wang W, Hann SS. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019;163:58-72. [Crossref] [PubMed]
- Yue B, Cai D, Liu C, et al. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther 2016;24:2064-77. [Crossref] [PubMed]
- Yu X, Mi L, Dong J, et al. Long intergenic non-protein-coding RNA 1567 (LINC01567) acts as a "sponge" against microRNA-93 in regulating the proliferation and tumorigenesis of human colon cancer stem cells. BMC Cancer. 2017;17:716. [Crossref] [PubMed]
- Luo J, Guo Y, Liu X, et al. Long non-coding RNA LINC01410 promotes colon cancer cell proliferation and invasion by inhibiting miR-3128. Exp Ther Med 2018;16:4824-30. [PubMed]
- Zhou P, Sun L, Liu D, et al. Long Non-Coding RNA lincRNA-ROR Promotes the Progression of Colon Cancer and Holds Prognostic Value by Associating with miR-145. Pathol Oncol Res 2016;22:733-40. [Crossref] [PubMed]


