
Multiple roles of THY1 in gastric cancer based on data mining
Introduction
According to the results of cancer statistics in 2019, the global incidence of gastric cancer (GC) ranked fifth, and mortality ranked third, second only to lung cancer and colorectal cancer (1). At present, radical gastrectomy is still the main method for the treatment of GC, but most of the patients undergoing surgery are diagnosed in the advanced stage, and the 5-year survival rate is very low. Although many oncogenes and tumor suppressor genes have been identified to play a vital role in the development of GC, there are still no specific cancer-targeted drugs for large-scale clinical applications. Therefore, exploring the molecular mechanism of GC development and finding reliable biomarkers as early diagnosis basis and reliable therapeutic targets have important clinical value for prolonging the survival of patients with GC.
THY1 is a GPI-anchored cell surface protein involved in cell-cell, cell-matrix interactions and is associated with a variety of cancers. In many cases, the expression of THY1 confirms the cancer stem cells and its over-expression in the tumor micro-environment enhances its ability of proliferation and metastasis (2-4). In others, the expression of THY1 inhibits tumor invasion and anchors independent growth (5). There are still many unresolved problems and challenges of THY1 related to the use as biomarker or drug target. Currently, little work has been carried on the mechanism or function of THY1 in GC. Therefore, it is meaningful to clarify the effect of THY1 in its development and progression.
Autophagy maintained cellular homeostasis by degrading dysfunctional or unnecessary proteins and organelles. Autophagy had been recognized as a cytoprotective process against environmental stress, such as starvation, ER stress, hypoxia, and pathogen infection (6,7). Autophagy is regulated by many signal pathways. For example, mTOR-dependent pathways which including AMPK/mTOR and PI3K/Akt/mTOR pathways (8,9). A growing number of studies reveal the critical role of autophagy in the development of GC (10,11), clarifying the autophagy-related mechanisms which would help to increase the therapeutic efficiency for GC. The present study focuses on the identification, clinical relevance and brief mechanism of THY1 in GC. Via data mining and analysis, we found that THY1 was over-expressed in GC tissues compared to normal ones. Importantly, results from the GEPIA and the Kaplan-Meier Plotter demonstrated that the expression level of THY1 was positively correlated to relapse, and negatively correlated to shorter OS, FP, and PPS. Consequently, we performed loss-of-function experiments after confirming obvious THY1 expression in GC cells and results showed that knockdown of THY1 restrained proliferation and migration of GC cells. Most importantly, it is found that THY1 knockdown can increase formation of autophagosomes and autophagic flux, indicating that THY1 expression levels affect GC cell autophagy. To further investigate the potential mechanism, we used various online databases. The outcome revealed that THY1 may be involved in cell-cell junction organization, Notch pathway, as well as immune regulation.
Methods
Ethics statement
All experiments using animal and human samples were reviewed and approved by the Ethics Committee of Nanjing Medical University (No. 2019669). Written informed consent for experimental use of the specimens was acquired from each participant, and the research was carried out according to the relevant guidelines and regulations.
Tools for THY1 expression level analysis
TCGA portal (www.tcgaportal.org) and FIREBROWSE (http://firebrowse.org/) were applied to detect the expression of THY1 in diverse tumor tissues and corresponding para-carcinoma tissues. The Human Protein Atlas (https://www.proteinatlas.org/), which compiles many reports and forms of the tissues, cells and pathology atlas, as well as gene information in tissues and cells were applied to explore the expression of THY1 in human tissues and location of THY1 mRNA in cells.
Tool for genes correlations analysis
GEPIA2 (http://gepia2.cancer-pku.cn/#index) is a dataset available for analyzing the RNA sequencing resources from the TCGA and the GTEx projects, containing 9,736 tumors and 8,587 normal samples. Customizable functions are provided such as similar gene detection, tumor/normal differential expression, patient survival curve, profiling based on tumor types or pathological stages, correlation analysis and dimensionality reduction analysis. It was utilized through the whole research to assess the correlations between all vital genes.
Tools for protein-protein interaction and functional enrichment analysis
Metascape (http://metascape.org/gp/index.html#/main/step1) is a web-based portal designed to leverage more than 40 independent knowledge bases, which combines gene annotation, interactome analysis, functional enrichment and membership search. Heatmap and network of enrichment terms associated with THY1 were acquired from Metascape. STRING (https://string-db.org/cgi/input.pl) is a database of known and predicted interactions between proteins which include associations directly (physically) or indirectly (functionally). They are originated from computational predictions, transferred knowledge between organisms and interactions which generated from other databases. We used STRING to produce an interaction network between THY1 and critical proteins.
Tool for immune-related analysis
DISIDB (http://cis.hku.hk/TISIDB/index.php) is a web valuable for tumor and immune system interaction, which integrates various heterogeneous data types. It was applied to analyze the spearman correlation between expression of THY1 and immunomodulators.
Cells culture and transfection
GC cells was cultured in RPMI-1640 medium (Biological Industries, Israel) containing 10% fetal bovine serum (Biological Industries, Israel) and 1% Penicillin-Streptomycin (Beyotime, China) at 37 °C in a humidified incubator with 5% CO2. Non-targeting control siRNA (si-NC) and small interfering RNA against THY1 (Si-THY1) and were synthesized by Guangzhou RiboBio Company. Transfections were conducted using the Opti-MEM (GiGCo, USA) and Lipofectamine 2000 (Invitrogen, USA). 48 h post-transfection, Cells were harvested for further study. The target sequence of si-THY1was shown in Table 1.
Table 1
| Name | Target sequence |
|---|---|
| si-RNA1 | 5'-CAGCCUGACCCGUGAGACATT-3' |
| si-RNA2 | 5'-GAACCAACUUCACCAGCAATT-3' |
| si-RNA3 | 5'-ACAAACUGGUCAAGUGUGATT-3' |
| THY1 | 5'-GTTTGACCAGGAAAGCAGCG-3' (F) |
| 5'-CTCTTGGGAGCTTGGGACAG-3' (R) | |
| GAPDH | 5'-TCGACAGTCAGCCGCATCTTCTTT-3' (F) |
| 5'-ACCAAATCCGTTGACTCCGACCTT-3' (R) |
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted by RNA extraction kit (TIANGEN, China) following the manufacturer’s instruction. QRT-PCR was conducted to detect the expressions of genes using the Master Mix kit (YEASHEN, China). GAPDH was used for internal control. The outcomes were evaluated via the −ΔCt method. The primer pair for THY1 was shown in Table 1.
Cell proliferation experiments
Cell proliferation was measured with the Cell Counting Kit-8 (CCK-8). Firstly, the cells were transfected with either Si-THY1 or si-NC and cultured at 37 °C. Then the CCK-8 solution (Dojindo, Japan) was added into each well following incubation for 2 h. The absorbance was measured on 0, 24, 48, 72 and 96 h time points at a wavelength of 450 nm. All experiments were conducted three more times.
Wound healing assay
Cells were suspended and seeded in 6-well plates after transfection with 100 nm si-THY1 or si-NC. Artificial linear wounds were made with a standard 200 µL pipette tip on the fused cell monolayer. Free floating cells were removed by PBS washing and the medium was added to the plate for further incubation. The width of the scratch gap was examined using an inverted microscope and imaged at 0 and 12 hours. Each experiment was performed in triplicate due to the difference in the width of the original wound and the quantitative cell migration.
LysoTracker red staining
The cells were transfected with si-NC or 100 nm si-THY1, and cultured in 12-well plates at 60–70% density following 24 h incubation. After that, cells were washed twice by PBS and stained with 50 nM of lysoTracker Red DND-99 (Invitrogen, USA) protected from light for 30 min and then examined using a confocal laser scanning microscope.
mCherry-enhanced green fluorescent protein (EGFP)-light chain (LC)-3B transfection and imaging
The cells were counted and seeded in 12-well plates in complete culture medium one day prior to transfection. Transfection with lentiviral mCherry-EGFP-LC3B (GenePharmaCo, China) was conducted according to the manufacturer’s instructions. Specific clones were selected with puromycin and identified using a flow cytometer. Subsequently, the cells were seeded onto 12-well plates for attachment overnight; following transfection with 100 nm Si-THY1 or si-NC, the cellular punctate pattern of mCherry-EGFP-LC3B was detected using a confocal microscope.
Western blot assay
The total proteins were extracted and the protein concentration was measured respectively. Then, the proteins were loaded onto a gel, SDS-PAGE was performed, and then proteins were transferred onto PVDF membranes (EMD Millipore, USA). After blocking for 2 h with 5% skim milk at 4 °C, the membranes were incubated overnight with the primary antibodies on a shaking table at 4 °C. The primary antibodies used were as follows: anti-p62 (1:10,000; Abcam, UK), anti-LC3B (1:2,000; Sigma-Aldrich, USA) and GAPDH (1:10,000; Abcam, UK). Then the membranes were washed three times and blocked with a HRP-conjugated secondary antibody (1:5,000; ABclonal, China) for a further 2 h at 4 °C. Visualization was conducted using an ECL kit (Beyotime, China).
Statistical analysis
Data are showed as mean ± standard deviation (SD). All statistical analysis was evaluated by GraphPad Prism 7 with the Student’s t test or one-way analysis of variance ANOVA. The correlations of gene expressions were evaluated by Spearman’s correlation. P<0.05 was considered as statistically significant.
Results
THY1 is over-expressed in GC and has remarkable clinical significance
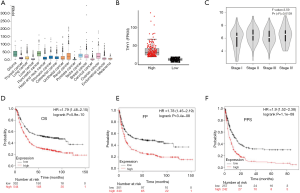
We initially explored the THY1 expression in normal tissues and GC tissues using TGCA. The results showed that THY1 has expression in most human cancers (Figure 1A) and has significant higher expression in GC tissues than that in normal ones (Figure 1B). GEPIA showed that THY1 expression is positively associated with the TNM stage of GC patients (Figure 1C). To further confirm the prognostic potential of THY1 in GC, Kaplan-Meier Plotter were used. As presented in Figure 1D,E,F, that higher THY1 expression is associated with shorter OS, FP and PPS compared to lower expression.
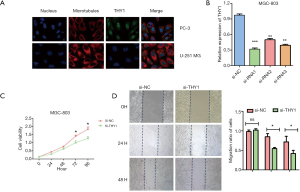
Knockdown of THY1 suppresses proliferation and migration of GC cells
The Human Protein Atlas database showed that in PC-3 and U-251 MG cell lines, THY1 located in the nucleus and membrane (Figure 2A). To explore the physiological characteristics role of THY1 in tumor, CCK8 assay and wound healing assay was conducted in MGC-803 cell line. As shown in our results, THY1 was significantly reduced using the Si-THY1 (Figure 2B). Knockdown of THY1 could inhibit cell proliferation compared to vehicle group, indicating the inhibiting role of THY1 knockdown on GC cells aggression (Figure 2C). Moreover, the suppression of THY1 by Si-THY1 showed a lower scratch closure rate or relative migration rate compared to controlled ones seen from wound healing assay (Figure 2D). These results suggest that inhibition of THY1 could undermine the progression of GC in vitro including proliferation and metastasis.
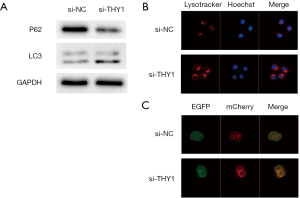
THY1 expression levels affect GC cell autophagy
Autophagy has a dual role in GC, including a tumour-suppressor role and a tumour-promoter role. In this study, we examined the relationship between THY1 and autophagy-related protein expression in GC cells that were transfected with si-THY1. LC3 and P62 are viewed as autophagy-related markers with GC prognostic value. We found that the expression levels of LC3B increased most markedly after THY1was down-expressed for 24 h, while P62 protein expression was significantly decreased (Figure 3A). When visualized by immunofluorescence, the acidotropic dye probe, LysoTracker Red, was used to label and track cellular acidic organelles, including lysosomes and autolysosomes in living cells. As presented in Figure 3B, the red fluorescence was much brighter in cells transfected with Si-THY1for 24 h. In addition, dual-fluorescent cancer cell lines were constructed via mCherry-EGFP-LC3B transfection to detect autophagic flux in GC cells. The Red (mCherry) and green (EGFP) fluorescence indicated the formation of autophagosomes. As shown in Figure 3C, it is evident that Si-THY1transfection resulted in increased levels of fluorescence. Therefore, these data suggest that THY1 knockdown induce extensive autophagic flux in GC cell.
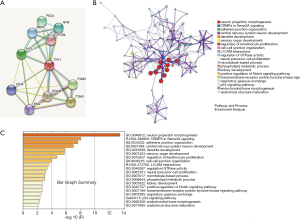
Enrichment analysis and interactive proteins network
The interaction network analysis from STRING displayed some proteins including CD34, ALPL, ITGB3, et al., which could bind directly with THY1 (Figure 4A). Metascape was used to perform the enrichment analysis of co-expression genes and the results showed that THY1is mainly involved in the processes of cell-cell junction, L1CAM interactions, phosphorylation process, and p53 signal pathway (Figure 4B,C).
THY1 expression is correlated with immune system
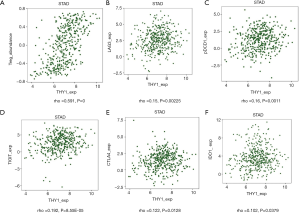
More and more researches have proved that immune system is highly related to tumor process. We investigated the relationship between THY1 expression and immune factors. Results showed that THY1 was significantly linked with Tregs (Figure 5A). Some Immunosuppressive membrane proteins whose expressions including LAG3, PDCD1, TIGIT, CTLA4, IDO1 have significant correlation with THY1expression by filtering: P<0.05 as well as |±rho| ≥0.1 were listed (Figure 5B,C,D,E,F).
Discussion
Nowadays, the application of effective tumor biomarkers has become one of hot issues in the field of cancer research, and is associated with the patient’s quality of life and survival outcomes (12,13). THY1 has been identified over-expressed on the cytoplasmic membrane in several cell types. Human THY1 gene is located on the long arm of chromosome 11 (11q23.3) (14). It activates a serious of cellular events, such as, proliferation, differentiation, wound repair and apoptosis, which makes it a valuable diagnostic marker exhibiting prognostic significance. In the current study, we found that THY1 is over-expressed in GC compare with normal tissues and is correlated with TNM stage, OS, FP and PPS. Besides, to evaluate whether THY1-silencing contribute to anti-cancer effect in GC, in vitro studies were conducted, which confirmed THY1 knockdown could inhibit GC cell proliferation and migration in a time-dependent manner, indicating it could be a potential bio-marker in GC.
Previous studies show THY1 potent to be a marker to confirm cancer stem cells in GC and differentiated acute myeloid leukemia (AML) subtypes, that, as THY1 was highly expressed in cancer stem cells which expressed self-renewal or drug-resistance genes to maintain and reinforce their stemness (15,16). In human ovarian cancer, THY1 has been linked to tumor suppression (17). It promotes the metastasis of human melanoma through mediating the adhesion of melanoma cells to endothelial cells (18). Kawamura et al. reported THY1 differentiated between malignant pleural mesothelioma and lung carcinoma in lung cancer (19,20). The expression of THY1 has also contributed to poor survival in hepatocellular carcinoma (21). Besides, Cheng et al. showed that exogenous overexpression of THY1 led to the upregulation of Cyclin D1, down regulation of E-cadherin which resulted in hepatocarcinoma progression and metastasis (22). The overexpression of THY1 was associated with increased lymph node metastasis in esophageal squamous carcinoma cells. Moreover, THY1+ cells were found to be resistant to taxotere treatment (23). Thus, THY1+ cells have a great potential in targeting cancer stem cells and suppressing their metastasis in hepatocellular carcinoma and esophageal carcinoma.
Autophagy is an evolutionarily conserved, lysosome-mediated degradative process (24), whereby intracellular organelles and macromolecules are recycled by the cell. It plays a crucial role in sustaining the homeostasis of cells and various malignant diseases. Moreover, autophagy is quite complex in malignant tumors (25,26). It is thought that autophagy prevents tumor genesis; however, the increased autophagic flux usually promotes cancer cell growth following tumor formation (27,28). In addition, it has also been reported that autophagy is involved in the anti-tumor catabolism of tumor genesis (29). Autophagy-associated genes serve an important role during autophagy, of which, LC3II serves as a key molecular marker of autophagy (30). As a selective substrate for autophagy, P62 undergoes hydrolysis during autophagy (31). Therefore, LC3 and P62 are the most commonly used tools for detecting autophagy. Our results revealed that with THY1 knockdown, the levels of LC3II expression increased, and the expression of P62 decreased, indicating the enhanced level of autophagy. Moreover, the induction of autophagy was observed directly using a laser confocal microscope. Following THY1 knockdown, the number of autophagolysosomes and autophagosomes were highly increased as determined by lysotracker staining and mCherry-EGFP-LC3B transfection, respectively. However, we did not know whether the THY1 knockdown induced autophagy by directly mechanisms, the specific mechanism needs to be further verified.
Tregs are part of the CD4+ T cell population. They overexpress the transcription factor forkhead box P3 (FoxP3) and CD25, and are capable of suppressing immune responses (32,33). Our prediction showed that THY1expression is highly associations with Tregs and immune markers including lymphocyte activation gene-3 (LAG3), programmed cell death 1 (PDCD1), as the immune co-inhibitory receptors LAG3 and PDCD1 contribute to tumor evasion and autoimmunity synergistically. Fortunately, monoclonal antibodies together with protein toxins have been evaluated in vivo models of leukemia. Previously, Ramakrishnan et al. reported using an anti-THY-1.1antibody disulfide along with pokeweed antiviral protein (PAP) and ricin A chain, which suppress synthesis of protein via blocking the 60S ribosomal subunit, to counteract cancer cell development (34). It is important to consider therapy targeting THY1 with the increasing understanding of THY1 in cancer cells and tumor microenvironment. Further researches in exploring the certain regulation of different cancer types and expression of THY1 will be helpful for achieving better therapeutic strategies.
Conclusions
The over-expression of THY1 is correlated with GC progression and prognosis. Monitoring the expression of THY1 is conducive to the timely assessment of GC development. THY1 expression levels also affect cellular autophagy, which suggested that autophagy regulation could become a promising approach to enhance the GC therapy efficiency. In addition, THY1 can be used as a marker molecule for evaluating the tumor microenvironment status of GC patients and a target for immunotherapy.
Acknowledgments
We thank Betty Zhang (Michael G. DeGroote School of Medicine, McMaster University, Hamilton, Ontario, Canada) for her linguistic assistance during the preparation of this manuscript.
Funding: This research was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.51). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments using animal and human samples were reviewed and approved by the Ethics Committee of Nanjing Medical University (No. 2019669). Written informed consent for experimental use of the specimens was acquired from each participant, and the research was carried out according to the relevant guidelines and regulations. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69:7-34. [Crossref] [PubMed]
- Chen WC, Chang YS, Hsu HP, et al. Therapeutics targeting CD90-integrin-AMPK-CD133 signal axis in liver cancer. Oncotarget 2015;6:42923-37. [Crossref] [PubMed]
- Lobba AR, Forni MF, Carreira AC, et al. Differential expression of CD90 and CD14 stem cell markers in malignant breast cancer cell lines. Cytometry A 2012;81:1084-91. [Crossref] [PubMed]
- Zhang K, Che S, Pan C, et al. The SHH/Gli axis regulates CD90-mediated liver cancer stem cell function by activating the IL6/JAK2 pathway. J Cell Mol Med 2018;22:3679-90. [Crossref] [PubMed]
- Cheng Y, Stanbridge E, Kong H, et al. A functional investigation of tumor suppressor gene activities in a nasopharyngeal carcinoma cell line HONE1 using a monochromosome transfer approach. Genes Chromosomes Cancer 2000;28:82-91. [Crossref] [PubMed]
- Galati S, Boni C, Gerra M, et al. Autophagy: A Player in response to Oxidative Stress and DNA Damage. Oxid Med Cell Longev 2019;2019:5692958. [Crossref] [PubMed]
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019;9:1167-81. [Crossref] [PubMed]
- Zhang Y, Fan Y, Huang S, et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci 2018;109:3865-73. [Crossref] [PubMed]
- Jiang X, Lu W, Shen X, et al. Repurposing sertraline sensitizes non-small cell lung cancer cells to erlotinib by inducing autophagy. JCI Insight 2018; [Crossref] [PubMed]
- Masuda GO, Yashiro M, Kitayama K, et al. Clinicopathological Correlations of Autophagy-related Proteins LC3, Beclin 1 and p62 in Gastric Cancer. Anticancer Res 2016;36:129-36. [PubMed]
- Zhang HQ, He B, Fang N, et al. Autophagy inhibition sensitizes cisplatin cytotoxicity in human gastric cancer cell line SGC7901. Asian Pac J Cancer Prev 2013;14:4685-8. [Crossref] [PubMed]
- Sun Z, Jia J, Du F, et al. Clinical significance of serum tumor markers for advanced gastric cancer with the first-line chemotherapy. Transl Cancer Res 2019;8:2680-90. [Crossref]
- Wu G, Li J, Qin C. Reduced RANBP9 expression is associated with poor prognosis in colorectal cancer patients. Transl Cancer Res 2019;8:2704-12. [Crossref]
- Seki T, Spurr N, Obata F, et al. The human Thy-1 gene: structure and chromosomal location. Proc Natl Acad Sci U S A 1985;82:6657-61. [Crossref] [PubMed]
- Notta F, Doulatov S, Laurenti E, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 2011;333:218-21. [Crossref] [PubMed]
- Shaikh MV, Kala M, Nivsarkar M. CD90 a potential cancer stem cell marker and a therapeutic target. Cancer Biomark 2016;16:301-7. [Crossref] [PubMed]
- Chen WC, Hsu HP, Li CY, et al. Cancer stem cell marker CD90 inhibits ovarian cancer formation via β3 integrin. Int J Oncol 2016;49:1881-9. [Crossref] [PubMed]
- Schubert K, Gutknecht D, Köberle M, et al. Melanoma cells use Thy-1 (CD90) on endothelial cells for metastasis formation. Am J Pathol 2013;182:266-76. [Crossref] [PubMed]
- Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One 2008;3:e2637. [Crossref] [PubMed]
- Kawamura K, Hiroshima K, Suzuki T, et al. CD90 is a diagnostic marker to differentiate between malignant pleural mesothelioma and lung carcinoma with immunohistochemistry. Am J Clin Pathol 2013;140:544-9. [Crossref] [PubMed]
- Lingala S, Cui Y, Chen X, et al. Immunohistochemical staining of cancer stem cell markers in hepatocellular carcinoma. Exp Mol Pathol 2010;89:27-35. [Crossref] [PubMed]
- Cheng BQ, Jiang Y, Li DL, et al. Up-regulation of thy-1 promotes invasion and metastasis of hepatocarcinomas. Asian Pac J Cancer Prev 2012;13:1349-53. [Crossref] [PubMed]
- Tang KH, Dai YD, Tong M, et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res 2013;73:2322-32. [Crossref] [PubMed]
- Hurley JH, Young LN. Mechanisms of Autophagy Initiation. Annu Rev Biochem 2017;86:225-44. [Crossref] [PubMed]
- Russo M, Russo G. Autophagy inducers in cancer. Biochem Pharmacol 2018;153:51-61. [Crossref] [PubMed]
- Lin JF, Hwang TI. Autophagy regulation in bladder cancer as the novel therapeutic strategy. Transl Cancer Res 2017;6:S708-S719. [Crossref]
- Amaravadi R, Kimmelman A, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913-30. [Crossref] [PubMed]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012;12:401-10. [Crossref] [PubMed]
- Onorati AV, Dyczynski M, Ojha R, et al. Targeting autophagy in cancer. Cancer 2018;124:3307-18. [Crossref] [PubMed]
- Klionsky D, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1-222.
- East DA, Fagiani F, Crosby J, et al. PMI: a ΔΨm independent pharmacological regulator of mitophagy. Chem Biol 2014;21:1585-96. [Crossref] [PubMed]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 Programs the Development and Function of CD4+CD25+ Regulatory T Cells. J Immunol 2017;198:986-92. [PubMed]
- Thornton A, Shevach E. Tregs, Helios and tumor immunity: the sun has not yet risen. Transl Cancer Res 2016;5:S672-S4. [Crossref] [PubMed]
- Ramakrishnan S, Houston L. Prevention of growth of leukemia cells in mice by monoclonal antibodies directed against Thy 1.1 antigen disulfide linked to two ribosomal inhibitors: pokeweed antiviral protein or ricin A chain. Cancer Res 1984;44:1398-404. [PubMed]


