
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-2020-007
Metformin up-regulated miR-107 expression and enhanced the inhibitory effect of miR-107 on gastric cancer growth
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the third leading cause of cancer-related death worldwide, after lung and colorectal cancers (1). The pathogenesis of GC is a complex, multifactorial process involving epigenetic alterations as well as tumor gene mutations (2). Although the prognosis for individuals diagnosed with GC has improved with advances in treatment strategies, the overall patient survival rate is still very poor (3). Metformin is a widely used biguanide hypoglycemic agent that has been reported to decrease the risk of GC cancer progression (4). Previous studies have demonstrated that long-term metformin treatment in GC patients results in significantly improved disease-free survival and overall survival (OS) rates (4,5).
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play a vital role in various physiological and pathological processes by negatively regulating the stability or translational efficiency of their target mRNAs (6). Abnormal miRNA expression level has been observed in a variety of diseases, particularly in cancer. It has been shown that deregulation of miRNAs can lead to the progression and development of cancer, and can also be used to predict therapeutic response in cancer patients (7,8). MiR-107, which is found on chromosome 10, has an anti-oncogenic role and its expression is suppressed in multiple cancers including colorectal, breast, lung, and GCs (9-12). A previous study demonstrated that miR-107 over-expression inhibited proliferation and metastasis of GC cells, and that low miR-107 expression level may serve as an effective biomarker for predicting poor disease prognosis in GC patients (13).
Recent evidence shows that metformin exerts its anti-cancer effects through regulation of miRNAs (14-16). However, little is known about the effects of metformin on miR-107 in GC. In this study, we aimed to determine the effect of metformin treatment on miR-107 expression in GC and explore a novel, miR-107-driven molecular mechanism of metformin action in GC.
Methods
Chemicals and reagents
3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT), dimethyl sulfoxide (DMSO), streptomycin, penicillin, anhydrous alcohol, crystal violet, and Hoechst 33258 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Fetal bovine serum (FBS) and RPMI-1640 medium were purchased from HyClone (Logan, UT, USA) and Hangzhou Sijiqing Biological Engineering Materials Co., Ltd. (Hangzhou, China), respectively. Mimic-miR-107 and mimic-NC were obtained from Genechem (Shanghai, China).
Cell culture
The SGC-7901 and MGC803 cells were purchased from the China Center for Type Culture Collection (Wuhan, China) and cultured in (R0883, Sigma-Aldrich) (10% FBS, 100 U/mL streptomycin (15140122, Thermo Fisher Scientific) at 37 °C in a humidified atmosphere containing 5% CO2.
Data collection
The gene chip dataset (GSE30289) was obtained from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/), as described as previously (17). A custom microarray platform was used to analyze the expression levels of 985 human miRNAs in untreated and metformin-treated GC samples.
Tissue RNA extraction
The tissue was ground in liquid nitrogen and 1 mL of TRIzol (10296010, Thermo Fisher Scientific) was added for every 50–100 mg of tissue. Homogenization was performed with a homogenizer. Chloroform (0.2 mL) (496189, Sigma-Aldrich) was added to the TRIzol (1 mL), shaken vigorously for 15 s, and allowed to rest for 5 min at room temperature. The samples were centrifuged at 10,000 g for 15 min at 4 °C. The aqueous phase was transferred to a new 1.5 mL Eppendorf tube and an equal volume of isopropanol (34863, Sigma-aldrich) was added. The RNA pellets were washed by adding at least 1 mL of 75% ethanol (459844, Sigma-aldrich) for every 1 ml of TRIzol and were centrifuged at 7,500 g for no more than 5 min at 4 °C.
Quantitative real-time PCR and bioinformatics analysis
The analysis of miR-107 expression in different kinds of cancer in OncomiR (http://www.oncomir.org/) and the real-time qRT-PCR process were performed as previously reported (18,19). The specific miRNA primers were synthesized by Sangon Biotech (Shanghai, China). The primers for MAPK8 miRNA were 5'-TCTCTGGTCAAGTCAATGGTATG-3' and 5'-GACAATCAGCATCACAGTTCAC-3'. The primers for miR-107 were 5'-GCCCTGTACAATGCTGCT-3' and 5'-CAGTGCAGGGTCCGAGGTAT-3'. The primers for U6 were 5'-CTCGCTTCGGCAGCACATATACT-3' and 5'-ACGCTTCACGAATTTGCGTGTC-3'. The following protocol was used: 95 °C for 3 min, followed by 39 cycles of 95 °C for 10 s, followed by 60 °C for 30 s. The relative expression level of miR-107 was normalized to that of internal control U6 by using the 2−ΔΔCt cycle threshold method.
MTT assay
SGC-7901 and MGC803 cells were seeded at a density of 1×104 cells/mL on 96-well culture plates (3690, Corning, NYC, USA) with RPMI-1640 medium. Following incubation for 24 h at 37 °C, the cells were transfected with miR-107 mimics and treated with 20 mmol/L metformin (D150959-5G, Sigma-aldrich) for 24, 48, 72, or 96 h. Subsequently, 5 mg/mL MTT (10 µL) was added to each well and cells were cultured at 37 °C for 3 h. Then, DMSO (150 µL) (34869, Sigma-aldrich) was added to each well and the optical density was read at 490 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Colony formation assays
SGC-7901 and MC803 cells were seeded at 1×104 cells/well in a 6-well plate (657160, Corning, NYK, USA). After transfection with miR107 mimics, cells were treated with 20 mmol/L metformin for 72 h. After an additional 48 h of culture, cells were fixed in anhydrous alcohol (459844, Sigma-aldrich) for 15 min and stained with 0.1% crystal violet (29288, Sigma-aldrich) for 10 min. Finally, cells were washed three times using 0.05 mol/L phosphate buffered saline (PBS) (D8537, Sigma-aldrich) before being photographed and counted.
Immunoblotting
MNK28 and SGC-7901 cells were harvested. For immunoblotting, cells were lysed by RIPA buffer (P0013C, Beyotime) and centrifugated at 10,000 g for 10 min. Then extracted proteins were resolved by SDS-PAGE. The protein blots were incubated with appropriate antibodies and protein bands were visualized by ECL or ECLPLUS (Amersham, Piscataway, NJ, USA). The antibodies were used as follows: MAPK8 (#9252, Cell Signaling Technology, Boston, MA, USA), anti-Tubulin (#T8203, Sigma-Aldrich, St Louis, MO, USA). GeneGnome HR system (Syngene, Cambridge, UK) was used for blots scanned.
Construction of gene regulatory network
The publicly available databases transcriptome-wide microRNA target prediction (miRcode, http://www.mircode.org/) and TargetScan (http://www.targetscan.org/) were used to predict the target of miR-107. Cytoscape software was used to construct the regulatory network for metformin. Additionally, Venn (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to verify the common targets of miR-107 and metformin. Targetscan (http://www.targetscan.org/vert_71/) was used to predict the position of miR-107 binding to MAPK8.
Survival analysis
OS was calculated using a Kaplan-Meier (KM) curve. KM Plotter (http://kmplot.com/analysis/index.php?p=background) was used to evaluate the impact of 54,675 genes on the survival time of cancer patients. A total of 10,188 cancer samples were analyzed in KM Plotter, including 1,648 ovarian, 4,142 breast, 1,065 GC, and 2,437 lung sample microarray expression profiles. In this study, we analyzed the effect of miR-596 (high vs. low expression) on OS of GC patients, with a log rank P value and the hazard ratio (HR) using 95% confidence intervals (CI).
Kaplan-Meier Plotter online database was used to evaluate the prognostic value of miR-107 [high (n=229) vs. low (n=202) expression] and MAPK8 [high (n=298) vs. low (n=587) expression] in GC patients as previously reported (20). The hazard ratio (HR) with 95% confidence intervals (CI) and logrank P value were calculated and shown on the KM survival plot.
Statistical analysis
Statistical analyses were carried out using SPSS 19.0 software (IBM, Armonk, NY, USA). Comparison of differences between groups was completed using Student’s t tests, and all data were presented as mean ± SD. A P value of less than 0.05 was considered to be statistically significant.
Results
Metformin increases miR-107 expression in GC
To explore which miRNA is regulated by metformin in the treatment of GC, we analyzed the miRNA microarray dataset (GSE30289) from the publicly available gene expression omnibus (GEO) database. In this study, a miRNA expression absolute fold change >2 and FDR value <0.01 was considered to be different. As shown in Figure 1A, a total of 34 miRNAs were identified, among which the expression level of miR-107 showed the most significant difference between untreated (control) and metformin-treated GC tissues. We then analyzed expression levels of miR-596 in GC tissues, as well as corresponding noncancerous mucosal tissues from the same patient (n=10 patients, Table 1), and found that the expression of miR-107 was lower in GC tissues when compared to corresponding noncancerous mucosal tissues (Figure 1B).
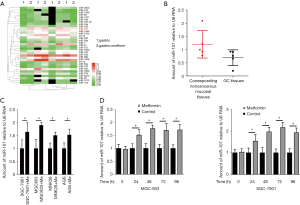
Table 1
| GC patients | Age (years) | Gender | Pathology | Tumor diameter (cm3) | Lymph node metastasis |
|---|---|---|---|---|---|
| Patient 1 | 59 | Male | Adenocarcinoma | Chemo-therapy | No |
| Patient 2 | 78 | Male | Adenocarcinoma | 3.7×3.5×1 | Yes |
| Patient 3 | 64 | Male | Adenocarcinoma | 3×2.7×0.6 | Yes |
| Patient 4 | 67 | Male | Adenocarcinoma | 4.5×3.5×1.5 | No |
| Patient 5 | 47 | Male | Adenocarcinoma | 8×7×2 | Yes |
| Patient 6 | 71 | Male | Adenocarcinoma | 4×2.5×1 | No |
| Patient 7 | 78 | Male | Adenocarcinoma | Chemo-therapy | Yes |
| Patient 8 | 60 | Female | Adenocarcinoma | 4×3.5×2 | No |
| Patient 9 | 58 | Male | Adenocarcinoma | 4×3.5×1 | Yes |
| Patient 10 | 75 | Female | Adenocarcinoma | 4.5×4×1.5 | Yes |
GC, gastric cancer.
We further examined miR-107 expression after treatment with metformin in several GC cell lines, including SGC-7901, MGC803, MNK28, and AGS, and found the expression of miR-107 to be highest in SGC-7901 and MGC803 (Figure 1C). Moreover, an RT-PCR assay was performed to validate miR-107 expression in SGC-7901 and MGC803 cells treated with metformin for 24, 48, 72, and 96 h compared to untreated (control) cells. As shown in Figure 1D, the expression of miR-107 was significantly upregulated with prolonged metformin treatment (P<0.05). These results suggest that metformin promotes the transcription of miR-107 in GC.
Metformin and miR-107 inhibit proliferation and invasion of SGC-7901 cells
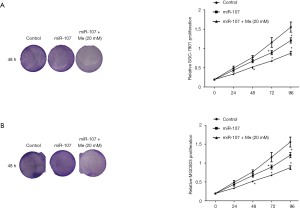
To investigate the effect of combined miR-107 expression and metformin treatment on the progression of GC, we transfected miR-107 mimics into metformin-treated SGC-7901 cells and performed gain-of-function assays in vitro. While SGC-7901 cell proliferation was only slightly decreased after transfection with a miR-107 mimic, combined treatment with a miR-107 mimic and metformin significantly decreased cell proliferation when compared to cells transfected with the NC mimic (P<0.05, Figure 2A). Similar results were obtained from colony formation assays and MTT assays in MGC803 cells (Figure 2B).
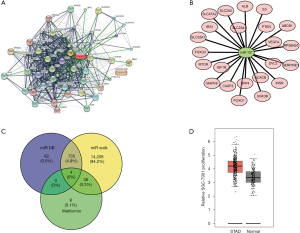
Bioinformatics analysis of regulatory network of miR-107 and metformin in GC
We performed further bioinformatics analyses to construct the gene regulatory network for metformin and miR-107. The targets of metformin and miR-107 were predicted by miRcode and TargetScan, and were then analyzed by Cytoscape software. We identified 50 metformin-mRNA regulatory relationships and 23 miR-107-mRNA regulatory relationships (Figure 3A,B). Moreover, Venn analysis showed a total of four genes that were predicted to be targeted by miR-107 and metformin simultaneously: caspase-3 precursor (CASP3), MAPK8, vascular endothelial growth factor (VEGFA) and ribosomal protein S6 kinase (RPS6KB1; Figure 3C). Based on these findings, we analyzed the Gene Expression Profiling Interactive Analysis (GEPIA) database and found that MAPK8 expression was markedly elevated in primary GC tissues compared to normal tissues (Figure 3D).
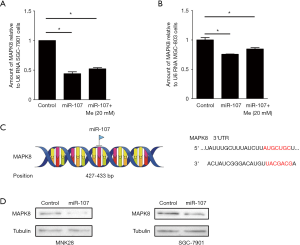
miR-107 down regulates the expression of MAPK8
To analyze MAPK8 mRNA expression, an RT-PCR assay was performed. After combined treatment with a miR-107 mimic and metformin, MAPK8 mRNA expression was significantly decreased in in SGC-7901 and MGC803 cells when compared with the control group (P<0.05; Figure 4A,B). Next, GEPIA was used to analyze the binding position of miR-107 on MAPK8. As shown in Figure 4B, miR-107 is predicted to bind to MAPK8 at 427-433 bp (Figure 4C). MAPK8 protein levels were slightly down-regulated after combined treatment with a miR-107 mimic and metformin (Figure 4D). Thus, these results indicate that MAPK8 acts as a target for miR-107 and metformin.
Target genes were bound up with survival of GC patients
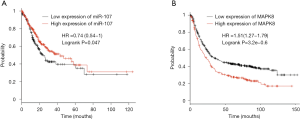
On the basis of the public Kaplan-Meier Plotter database, miR-107 and the MAPK8 were analyzed to verify their association with GC patient prognosis. As shown in Figure 5, patients with low expression of miR-107 had a significantly shorter survival probability (miR-107, P=0.047; Figure 5A). Conversely, patients with low expression of MAPK8 had significantly higher survival rates (P=3.2e-0.6; Figure 5B).
Discussion
Metformin has been the first choice for the treatment for type 2 diabetes since 1957, due in part to its low price and rare incidence of side effects. Numerous studies have reported that clinical usage of metformin is associated with a reduced GC risk (4,21). Sekino et al. showed that metformin suppresses cell proliferation and peritoneal metastasis in GC, and that the anti-tumor effect of metformin is enhanced in low-glucose conditions (22). Previous studies have also demonstrated that metformin achieves its anti-proliferative and anti-metastatic effects by mediating the expression of different miRNAs in multiple cancers, including breast, colorectal, cervical, and prostate cancers (15,16,23,24). However, there have only been a few relevant studies exploring this in GC. In the present study, we use bioinformatics analyses and qRT-PCR assays to demonstrate, for the first time, that metformin significantly increases miR-107 expression in the SGC-7901 GC cell line.
Previous studies have reported that nearly half of human miRNA genes are located in a tumor-associated mutation region, and aberrant miRNA expression may be strongly linked to the pathogenesis of various malignant tumors (25,26). MiR-107 has been shown to play a tumor-suppressive role by inhibiting tumor growth and invasion as well as inducing apoptosis and cell cycle arrest. Moreover, miR-107 expression is down-regulated in GC (9,27,28). Consistent with these previous reports, our study also found that miR-107 expression is significantly decreased in GC. Additionally, we found that an increased miR-107 expression combined with metformin treatment can further restrain the proliferative and invasive capacities of GC cells.
As the results of the bioinformatics analysis showed, CASP3, MAPK8, VEGFA, and RPS6KB1 were identified to be common target genes for miR-107 and metformin. These genes are also closely associated with infiltrating levels of CD8+ T cells and CD4+ T cells, as well as survival of GC patients. CASP3 is well described to be an important execution-phase caspase that is involved in cancer occurrence and progression (29). CASP3 was also reported to be a tumor suppressor in GC and its expression is correlated with a favorable prognosis in GC patients (30). In addition, MAPK8 is a member of the MAPK kinase family and is involved in many biological processes, including cell proliferation, differentiation, and transcription. Growing evidence has demonstrated that MAPK8 plays a crucial role in many cancers, including GC (31,32). VEGFA is as a member of the vascular endothelial growth factor family and is implicated in many cancer types; VEGFA tends to be up-regulated in cancer and is significantly correlated with tumor angiogenesis and metastasis (33). RPS6KB1 is reported to be an evolutionarily conserved serine/threonine kinase, and its alteration has the potential to be an important driver of tumor initiation and progression (34,35). However, more experiments, particularly in vivo studies, are necessary to elucidate further details about combined miR-107 and metformin treatment in GC.
Conclusions
Our results are the first to show that metformin observably enhances miR-107 expression and further strengthens the anti-proliferative and anti-metastatic role of miR-107 in GC. These results offer a novel therapeutic strategy of combined miR-107 and metformin treatment for GC patients.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.25). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by Ethical Committee of Zhejiang Cancer Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Abdi E, Latifi-Navid S, Zahri S, et al. Risk factors predisposing to cardia gastric adenocarcinoma: Insights and new perspectives. Cancer Med 2019;8:6114-26. [Crossref] [PubMed]
- Xia J, Shi D, Wu Z, et al. The poor prognosis of the primary gastric epithelioid angiosarcoma: A case report. Medicine (Baltimore) 2018;97:e0287. [Crossref] [PubMed]
- Courtois S, Lehours P, Bessede E. The therapeutic potential of metformin in gastric cancer. Gastric Cancer 2019;22:653-62. [Crossref] [PubMed]
- Lee CK, Jung M, Jung I, et al. Cumulative Metformin Use and Its Impact on Survival in Gastric Cancer Patients After Gastrectomy. Ann Surg 2016;263:96-102. [Crossref] [PubMed]
- Sun B, Liu C, Li H, et al. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol Lett 2020;19:595-605. [PubMed]
- Zhi F, Zhou G, Shao N, et al. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS One 2013;8:e72390. [Crossref] [PubMed]
- Cabello P, Pineda B, Tormo E, et al. The Antitumor Effect of Metformin Is Mediated by miR-26a in Breast Cancer. Int J Mol Sci 2016; [Crossref] [PubMed]
- Wang L, Li K, Wang C, et al. miR-107 regulates growth and metastasis of gastric cancer cells via activation of the PI3K-AKT signaling pathway by down-regulating FAT4. Cancer Med 2019;8:5264-73. [Crossref] [PubMed]
- Wang G, Ma C, Shi X, et al. miR-107 Enhances the Sensitivity of Breast Cancer Cells to Paclitaxel. Open Med (Wars) 2019;14:456-66. [Crossref] [PubMed]
- Fu Y, Lin L, Xia L. MiR-107 function as a tumor suppressor gene in colorectal cancer by targeting transferrin receptor 1. Cell Mol Biol Lett 2019;24:31. [Crossref] [PubMed]
- Lu C, Xie Z, Peng Q, et al. MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in non small cell lung cancer. Am J Cancer Res 2017;7:1863-73. [PubMed]
- Chen P, Zhao X, Wang H, et al. The Down-Regulation of lncRNA PCAT18 Promotes the Progression of Gastric Cancer via MiR-107/PTEN/PI3K/AKT Signaling Pathway. Onco Targets Ther 2019;12:11017-31. [Crossref] [PubMed]
- Ren W, Zhang X, Li W, et al. Exosomal miRNA-107 induces myeloid-derived suppressor cell expansion in gastric cancer. Cancer Manag Res 2019;11:4023-40. [Crossref] [PubMed]
- Sharma P, Kumar S. Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: pivotal role of superoxide dismutase (SOD). Cell Oncol (Dordr) 2018;41:637-50. [Crossref] [PubMed]
- Xia C, Liang S, He Z, et al. Metformin, a first-line drug for type 2 diabetes mellitus, disrupts the MALAT1/miR-142-3p sponge to decrease invasion and migration in cervical cancer cells. Eur J Pharmacol 2018;830:59-67. [Crossref] [PubMed]
- Kato K, Gong J, Iwama H, et al. The antidiabetic drug metformin inhibits gastric cancer cell proliferation in vitro and in vivo. Mol Cancer Ther 2012;11:549-60. [Crossref] [PubMed]
- Sarver AL, Sarver AE, Yuan C, et al. OMCD: OncomiR Cancer Database. BMC Cancer 2018;18:1223. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Li P, Zhang C, Gao P, et al. Metformin use and its effect on gastric cancer in patients with type 2 diabetes: A systematic review of observational studies. Oncol Lett 2018;15:1191-9. [PubMed]
- Sekino N, Kano M, Matsumoto Y, et al. The Antitumor Effects of Metformin on Gastric Cancer In Vitro and on Peritoneal Metastasis. Anticancer Res 2018;38:6263-9. [Crossref] [PubMed]
- Zhang J, Li G, Chen Y, et al. Metformin Inhibits Tumorigenesis and Tumor Growth of Breast Cancer Cells by Upregulating miR-200c but Downregulating AKT2 Expression. J Cancer 2017;8:1849-64. [Crossref] [PubMed]
- Wang Y, Wu Z, Hu L. The regulatory effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in the epithelial-mesenchymal transition(EMT) for colorectal cancer(CRC). Eur J Pharmacol 2018;834:45-53. [Crossref] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [Crossref] [PubMed]
- Zhao W, Zhang X, Liu J, et al. miR-27a-mediated antiproliferative effects of metformin on the breast cancer cell line MCF-7. Oncol Rep 2016;36:3691-9. [Crossref] [PubMed]
- Feng L, Xie Y, Zhang H, et al. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol 2012;29:856-63. [Crossref] [PubMed]
- Wang S, Ma G, Zhu H, et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci Rep 2016;6:36531. [Crossref] [PubMed]
- Kuo WT, Shen L, Zuo L, et al. Inflammation-induced Occludin Downregulation Limits Epithelial Apoptosis by Suppressing Caspase-3 Expression. Gastroenterology 2019;157:1323-37. [Crossref] [PubMed]
- Huang KH, Fang WL, Li AF, et al. Caspase-3, a key apoptotic protein, as a prognostic marker in gastric cancer after curative surgery. Int J Surg 2018;52:258-63. [Crossref] [PubMed]
- Li S, Liu R, Xue M, et al. Spleen tyrosine kinase-induced JNK-dependent NLRP3 activation is involved in diabetic cardiomyopathy. Int J Mol Med 2019;43:2481-90. [PubMed]
- Liu J, Wang T, Creighton CJ. JNK1/2 represses Lkb1-deficiency-induced lung squamous cell carcinoma progression. Nat Commun 2019;10:2148. [Crossref] [PubMed]
- Tamma R, Annese T, Ruggieri S, et al. VEGFA and VEGFR2 RNAscope determination in gastric cancer. J Mol Histol 2018;49:429-35. [Crossref] [PubMed]
- Wang-Bishop L, Chen Z, Gomaa A, et al. Inhibition of AURKA Reduces Proliferation and Survival of Gastrointestinal Cancer Cells With Activated KRAS by Preventing Activation of RPS6KB1. Gastroenterology 2019;156:662-675.e7. [Crossref] [PubMed]
- Hussain SS, Huang SB, Bedolla RG, et al. Suppression of ribosomal protein RPS6KB1 by Nexrutine increases sensitivity of prostate tumors to radiation. Cancer Lett 2018;433:232-41. [Crossref] [PubMed]


