
Construction of differentially expressed Her-2 related lncRNA-mRNA-miRNA ceRNA network in Her-2 positive breast cancer
Introduction
Breast cancer as the most common malignant tumor is the primary cause of cancer-related death among women worldwide. Breast cancer is a multifaceted and heterogeneous disease with different molecular subtypes (1). Base on the 2013 St Gallen classification of intrinsic breast cancer subtypes, breast cancer is classified into luminal A, luminal B, Her-2 enrich and triple negative breast cancer, according to estrogen receptor (ER), progesterone receptor (PR) and Her-2 status (2). The epidermal growth factor receptor (Her/Neu) family are composed of Her-1, Her-2, Her-3, and HER-4 which involved in the regulation of proliferation, migration and differentiation (3,4). Her-2 enrich subtype breast cancer is characterized as Her-2 gene amplification with increased invasiveness and aggressiveness accounting for 20–30% of invasive infiltrate breast cancer (5,6). Despite systemic anti-Her-2 therapy including novel monoclonal antibodies (trastuzumab and pertuzumab), small molecule inhibitors (lapatinib), significantly improve the outcome of Her-2 positive breast cancer, treatment resistance and potential cardiotoxicity still remain unsolved issues (7-10).
MicroRNAs (miRNAs) are small non-coding RNAs participate in several biological processes and play an important part to regulate mRNA expression (11,12). lncRNAs are non-coding RNAs (ncRNAs) ranging from 200 nucleotides to 100 kb in length, regulating the activity of mRNAs by direct interact with one or more miRNA response elements (MREs) (13,14). Many researches reveal that lncRNAs may play an important part in post-transcriptional regulation and transcriptional regulation, and represent potential early diagnosis biomarkers and therapeutic targets (15,16).
In 2014, Yang et al. present that upregulated FOXO1 and E-cardherin expression level in breast cancer cells inhibited EMT (epithelial–mesenchymal transition) and metastasis progress in breast cancer while miR-9 inactivating (17,18). The noncoding RNA CXCR4 sponging of miR-146a which may result the upregulated of expression level of TNF receptor-associated factor 6 (TRAF6) and epidermal growth factor receptor (EGFR), stimulate tumor cell proliferation, invasion and migration in breast carcinoma and other various cancer types (19-22).
Nonetheless, the molecular pathogenesis mechanism of ceRNA in Her-2 enrich subtype breast cancer have not been further investigated and discovered. It is important to explore interaction of miRNA and ceRNA which stable and differentially expressed in different breast cancer molecular subtypes, which may act as new molecular biomarkers
Methods
RNA expression data and clinical feature associations
RNA expression data sets and detailed clinical data of 1,109 invasive infiltrating ductal breast carcinoma tissues and 113 adjacent tissues were collected from TCGA database. The corresponding information met the inclusion criteria as follows: (I) histologic diagnosis were invasive infiltrating ductal breast cancer; (II) clinical data (TNM stage, age, gender, overall survival and ER, PR and Her-2 receptor) were complete. As result, 1,109 breast cancer samples were classified into two cohorts including ER+/PR+ cohort (n=461) and ER-/PR- cohort (n=152). Moreover, ER+/PR+ cohort were divided into ER+/PR+/Her-2+ group (n=98) and ER+/PR+/Her-2- group (n=39); and ER-/PR- cohort were divided into ER-/PR-/Her-2+ group (n=39) and ER-/PR-/Her-2- group (n=113).
Screening for DemRNAs, DemiRNAs, and DelncRNAs
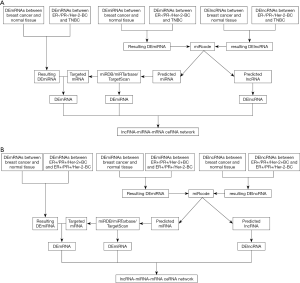
Differently expressed mRNAs, lncRNAs and miRNAs between 1,109 breast cancer samples and 113 normal samples were analysis by a Bioconductor package, edgeR based on R language with the threshold of |log2FC| >1.5 and P value <0.05. In ER+/PR+ breast cancer cohort, differently expressed mRNAs, lncRNAs and miRNAs between ER+/PR+/Her-2+ breast cancer (n=98) and ER+/PR+/Her-2- breast cancer (n=363) were screened with the same threshold of |log2FC| >1.5, false discovery rate (FDR) <5%, and P value <0.05. In ER-/PR- breast cancer cohort, differently expressed mRNAs, lncRNAs and miRNAs between ER-/PR-/Her-2+ breast cancer (n=39) and ER-/PR-/Her-2- breast cancer (n=113) were screened with the same threshold. lncRNA-miRNA interactions were preformed to predicted and verified by miRcode. miRNA-mRNA interactions were selected to predict targeted mRNAs of miRNAs by miRanda, Targetscan and miRTarBase.
Construction of lncRNA-miRNA-mRNA ceRNA network
To be more accurate, DEmRNAs, DEmiRNAs and DElncRNAs (breast cancer vs. normal tissue) were intersected with DEmRNAs, DEmiRNAs and DElncRNAs in ER+/PR+ and ER-/PR- cohort, respectively. The resulting mRNAs were also intersected with targeted mRNAs predicted by miRNAs selected by lncRNA-miRNA interactions in miRode. ceRNA network was integrated of interaction among retained lncRNA, miRNA and mRNA. The construction of the network was visualized using the Cytoscape 3.4.0 software.
The flow chart in ER+/PR+ and ER-/PR- breast cancer cohort for the construction of ceRNA networks are illustrated (Figure 1A,B).
Results
At last, a total of 461 ER+/PR+ cohort including 98 Her-2-positive and 363 Her-2-negative breast cancers and a total of 152 ER-/PR- cohort were enrolled in our research (Table 1).
Table 1
| Clinical feature | ER+/PR+ cohort | ER-/PR- cohort | |||
|---|---|---|---|---|---|
| Her-2- | Her-2+ | Her-2- | Her-2+ | ||
| Age (years) | |||||
| >60 | 207 | 45 | 82 | 21 | |
| ≤60 | 156 | 53 | 31 | 16 | |
| Stage | |||||
| I | 72 | 11 | 19 | 2 | |
| II | 201 | 56 | 70 | 23 | |
| III | 80 | 29 | 16 | 10 | |
| IV | 3 | 1 | 2 | 1 | |
| Not available | 8 | 1 | 3 | 1 | |
| Margin | |||||
| Positive | 28 | 4 | 3 | 6 | |
| Negative | 309 | 81 | 100 | 31 | |
| Not available | 27 | 13 | 10 | 0 | |
| Pathological T | |||||
| T1 | 109 | 18 | 26 | 6 | |
| T2 | 192 | 63 | 71 | 26 | |
| T3 | 51 | 12 | 12 | 2 | |
| T4 | 10 | 5 | 4 | 3 | |
| Tx | 2 | 0 | 0 | 0 | |
| Pathological N | |||||
| N0 | 173 | 40 | 73 | 10 | |
| N1 | 122 | 37 | 24 | 14 | |
| N2 | 39 | 13 | 12 | 5 | |
| N3 | 24 | 8 | 4 | 6 | |
| Nx | 6 | 0 | 0 | 2 | |
| Pathological M | |||||
| M0 | 313 | 78 | 96 | 36 | |
| M1 | 3 | 1 | 2 | 1 | |
| Mx | 48 | 19 | 15 | 0 | |
DEmRNAs, DEmiRNAs and DElncRNAs in ER+/PR+ breast cancer cohort
Two hundred and sixty-nine DEmRNAs including 176 downregulated and 93 upregulated DEmRNAs were identified (98 Her-2-positive vs. 363 Her-2-negative breast cancers) in ER+/PR+ cohort after intersection with 3,201 DEmRNAs (breast cancer vs. normal tissues). Fifteen DEmiRNAs including 6 downregulated and 9 upregulated were identified (98 Her-2-positive vs. 363 Her-2-negative breast cancers) in ER+/PR+ cohort after intersection with 143 DEmiRNAs (breast cancer vs. normal tissues). One hundred and twenty-nine DElncRNAs including 71 downregulated and 58 upregulated (98 Her-2-positive vs. 363 Her-2-negative breast cancers) were identified in ER+/PR+ cohort after intersection with 1,772 DElncRNAs (breast cancer vs. normal tissues).
DEmRNAs, DEmiRNAs and DElncRNAs in ER-/PR- breast cancer cohort
After intersected with DEmRNAs, DEmiRNAs and DElncRNAs (breast cancer vs. normal tissues). Six hundred and ninety-three DEmRNAs including 431 downregulated and 262 upregulated DEmRNAs were retained for future analysis. Twenty-five DEmiRNAs (17 downregulated and 8 upregulated) and 364 DElncRNAs (223 downregulated and 141 upregulated) were retained for future analysis, respectively.
Prediction of DElncRNA–DEmiRNA and DEmiRNA–DEmRNA interactions
Four DElncRNA-DEmiRNA pairs were retained base on online software including 4 DElncRNAs and 1 DEmiRNA. target mRNA of DEmiRNA were marked by Targetscan, miRanda and miRTarBase prediction software and simultaneously intersected with 269 DEmRNAs in ER+/PR+ cohort online. Finally, 2 target genes were obtained.
In ER-/PR- cohort, 25 DElncRNA-DEmiRNA pairs were retained and target genes of 4 DEmiRNAs was also predicted. In result one target gene was selected for future analysis in ER-/PR- breast cancer cohort.
Construction of mRNA-miRNA-lncRNA ceRNA network in ER+/PR+ and ER-/PR- cohort
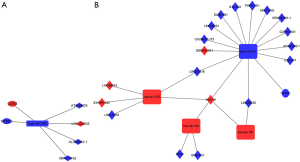
ceRNA network in ER+/PR+ breast cancer cohort was constructed of 4 DElncRNA–DEmiRNA pairs and 2 DEmiRNA–DEmRNA pairs, including 4 DElncRNAs, 1 DEmiRNAs, and 2 DEmRNAs. ceRNA network in ER-/PR- breast cancer cohort was constructed of 24 DElncRNA–DEmiRNA pairs and 1 DEmiRNA–DEmRNA pairs, including 19 DElncRNAs, 4 DEmiRNAs, and 1 DEmRNA. The ceRNA networks were visualized by Cytoscape software (Figure 2A,B).
Discussion
ncRNAs work as miRNA sponges in result to regulate miRNA activity and function. Additionally, many studies reveal that ncRNAs binding to miRNAs regulate many biological processes participated in proliferation and invasion of tumor cells (16,23). Previous studies reveal that Her-2 inhibition blockade PI3K-Akt-mTOR signal pathways, while PARP inhibitors showing anti-tumor activity in BRCA-related triple negative breast cancer (24).
The expression of Her-2 protein is regulated by several miRNAs, such as miR-34a, miR-31, miR15a, miR16, miR-363 and miR-21, which may modulate to increased multidrug resistance (25-28). In our study, ceRNA network is constructed to detect key RNAs in the progression and development of Her-2 enrich breast cancer downloaded from TCGA database. We divide breast cancer samples into different molecular subgroups according to gene expression–based classifications. MIR7-3HG- hsa-mir-204-NTRK2 axis is identified in both ER+/PR+ and ER-/PR- cohort in our study.
In previous studies reveal that downregulation of miR-204 accelerate breast cancer cell growth proliferation, and migration. Using real-time qPCR (RT-qPCR), Shen et al. show that compare to normal breast epithelial HBL-100 cells, the expression level of miR-204 is decreased in MCF-7 cells (29). Our analysis show that miR-204-5p is significantly downregulated in ER+/PR+/Her-2+ and ER+/PR+/Her-2- breast cancer cohort, which is also downregulated in ER-/PR-/Her-2+ and ER-/PR-/Her-2- breast cancer cohort. Neurotrophic tyrosine receptor kinase type 2 (NTRK2) as a novel target of miR-200cis aberrantly expressed in carcinoma cells (30). Common polymorphisms in NTRK2 influence the severity of symptoms and symptom burden in patients undergoing treatment for breast cancer (31). MIR7–3HG acts as a tumor suppressor, which promotes the expression of the tumor suppressor AMBRA1 (32). It is reported that modulating the expression of MIR7–3HG in lung cancer cells lead to a less proliferative state (33). However, in some studies MIR7–3HG overexpression is associated with poor prognosis in lung carcinoma (33). Gao et al. detect LINC00466-Has-mir-204-NTRK2 axis in invasive breast cancer which may associated with the prognosis of invasive breast cancer (34). Our result is consistent with previous studies which also present new therapeutic target for Her-2 positive breast cancer. In previous study, ZEB1 and CDH2 act as epithelial-to-mesenchymal transition (EMT) activators in breast cancer cells may inhibit the sensitivity of breast cancer cells to chemotherapy and enhance breast cancer cell progression and metastasis (35). We identify that CDH2 was upregulated ER+/PR+/Her-2+ subtype compared to ER+/PR+/Her-2- subtype, which may promote breast cancer cell EMT.
However, there are still some limitations in our study. Limited number of breast cancer patients are included in this study, and still need to be verified and confirmed in future studies. Other factors including races, stages, histological types and application of chemotherapy and endocrine therapy need to be accounted in. Predictive biomarkers to identify patients who will benefit from anti-Her-2 targeted therapy need to be verified in further analysis.
Conclusions
In conclusion, Her-2 related differently expressed lncRNAs, miRNAs and mRNAs are identified. According to ceRNA hypothesis, a potential Her-2 related regulatory ceRNA network may responsible for interpreting the mechanism underlying the biological processes of Her-2 positive breast cancer.
Acknowledgments
Funding: Research supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.34). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tella SH, Kommalapati A, Singhi RK. Cost-effectiveness in Managing Skeletal Related Events in Breast Cancer: A Strategy of Less-Intense Dosing Schedule of Bone Modifying Agents. Transl Cancer Res 2018;7:S81-4. [Crossref] [PubMed]
- Harbeck N, Thomssen C, Gnant M, St. . Gallen 2013: brief preliminary summary of the consensus discussion. Breast Care (Basel) 2013;8:102-9. [Crossref] [PubMed]
- Harbeck N. Advances in targeting Her-2-positive breast cancer. Curr Opin Obstet Gynecol 2018;30:55-9. [Crossref] [PubMed]
- Zhang B, Hurvitz S. Long-term outcomes of neoadjuvant treatment of Her-2-positive breast cancer. Clin Adv Hematol Oncol 2016;14:520-30. [PubMed]
- Asif HM, Sultana S, Ahmed S, et al. Her-2 Positive Breast Cancer - a Mini-Review. Asian Pac J Cancer Prev 2016;17:1609-15. [Crossref] [PubMed]
- Nagini S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anticancer Agents Med Chem 2017;17:152-63. [Crossref] [PubMed]
- Loibl S, Gianni L. Her-2-positive breast cancer. Lancet 2017;389:2415-29. [Crossref] [PubMed]
- Ahmed S, Sami A, Xiang J. Her-2-directed therapy: current treatment options for Her-2-positive breast cancer. Breast Cancer 2015;22:101-16. [Crossref] [PubMed]
- Singla H, Ludhiadch A, Kaur RP, et al. Recent advances in Her-2 positive breast cancer epigenetics: Susceptibility and therapeutic strategies. Eur J Med Chem 2017;142:316-27. [Crossref] [PubMed]
- Kast K, Schoffer O, Link T, et al. Trastuzumab and survival of patients with metastatic breast cancer. Arch Gynecol Obstet 2017;296:303-12. [Crossref] [PubMed]
- Dragomir M, Chen BQ, Calin GA. Exosomal lncRNAs as New Players in Cell-To-Cell Communication. Transl Cancer Res 2018;7:S243-52. [Crossref] [PubMed]
- Shah MY, Ferrajoli A, Sood AK, et al. microRNA Therapeutics in Cancer - An Emerging Concept. EBioMedicine 2016;12:34-42. [Crossref] [PubMed]
- Forrest ME, Khalil AM. Review: Regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett 2017;407:106-12. [Crossref] [PubMed]
- Achour C, Aguilo F. Long non-coding RNA and Polycomb: an intricate partnership in cancer biology. Front Biosci (Landmark Ed) 2018;23:2106-32. [Crossref] [PubMed]
- Jathar S, Kumar V, Srivastava J, et al. Technological Developments in lncRNA Biology. Adv Exp Med Biol 2017;1008:283-323. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol 2016;1402:271-86. [Crossref] [PubMed]
- Yang J, Li T, Gao C, et al. FOXO1 3'UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett 2014;588:3218-24. [Crossref] [PubMed]
- Liu DZ, Chang B, Li XD, et al. MicroRNA-9 promotes the proliferation, migration, and invasion of breast cancer cells via down-regulating FOXO1. Clin Transl Oncol 2017;19:1133-40. [Crossref] [PubMed]
- Chittasupho C, Anuchapreeda S, Sarisuta N. CXCR4 targeted dendrimer for anti-cancer drug delivery and breast cancer cell migration inhibition. Eur J Pharm Biopharm 2017;119:310-21. [Crossref] [PubMed]
- Fkih M’hamed I, Privat M, Trimeche M, et al. miR-10b, miR-26a, miR-146a and miR-153 Expression in Triple Negative Vs Non Triple Negative Breast Cancer: Potential Biomarkers. Pathol Oncol Res 2017;23:815-27. [Crossref] [PubMed]
- Shen H, Li L, Yang S, et al. Regulatory role of tumor necrosis factor receptor-associated factor 6 in breast cancer by activating the protein kinase B/glycogen synthase kinase 3β signaling pathway. Mol Med Rep 2017;16:2269-73. [Crossref] [PubMed]
- Papanastasiou AD, Sirinian C, Plakoula E, et al. RANK and EGFR in invasive breast carcinoma. Cancer Genet 2017;216-217:61-6. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Dey N, De P, Leyland JB. PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell signaling to clinical trials. Pharmacol Ther 2017;175:91-106. [Crossref] [PubMed]
- Liu C, Tong Z, Tan J, et al. MicroRNA-21-5p targeting PDCD4 suppresses apoptosis via regulating the PI3K/AKT/FOXO1 signaling pathway in tongue squamous cell carcinoma. Exp Ther Med 2019;18:3543-51. [PubMed]
- Kim D, Lee J, Kang J, et al. Notch1 in Tumor Microvascular Endothelial Cells and Tumoral miR-34a as Prognostic Markers in Locally Advanced Triple-Negative Breast Cancer. J Breast Cancer 2019;22:562-78. [Crossref] [PubMed]
- Yang K, Tang H, Ding M, et al. Expression of miR-195 and MEK1 in patients with bladder cancer and their relationship to prognosis. Int J Clin Exp Pathol 2019;12:843-50. [PubMed]
- Sun G, Sun L, Liu Y, et al. Her-2 expression regulated by downregulation of miR-9 and which affects chemotherapeutic effect in breast cancer. Cancer Gene Ther 2017;24:194-202. [Crossref] [PubMed]
- Shen SQ, Huang LS, Xiao XL, et al. miR-204 regulates the biological behavior of breast cancer MCF-7 cells by directly targeting FOXA1. Oncol Rep 2017;38:368-76. [Crossref] [PubMed]
- Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res 2011;13:R45. [Crossref] [PubMed]
- Young EE, Kelly DL, Shim I, et al. Variations in COMT and NTRK2 Influence Symptom Burden in Women Undergoing Breast Cancer Treatment. Biol Res Nurs 2017;19:318-28. [Crossref] [PubMed]
- Li Q, Wu X, Guo L, et al. MicroRNA-7-5p induces cell growth inhibition, cell cycle arrest and apoptosis by targeting PAK2 in non-small cell lung cancer. FEBS Open Bio 2019;9:1983-93. [Crossref] [PubMed]
- Capizzi M, Strappazzon F, Cianfanelli V, et al. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy 2017;13:554-66. [Crossref] [PubMed]
- Gao C, Li H, Zhuang J, et al. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. Cancer Manag Res 2018;11:1-11. [Crossref] [PubMed]
- Lee JW, Guan W, Han S, et al. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci 2018;109:1404-13. [Crossref] [PubMed]


