
Low expression of citron kinase is associated with poor patient outcomes in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common tumor type and the third leading cause of death related to cancer worldwide (1). Up to half of all HCC cases occur in China, and there were a total of 782,500 new liver cancer cases and 745,500 deaths worldwide in 2012, with 84.6% of liver cancer incidence and 86.3% of liver cancer deaths in the WHO Western Pacific region (WPRO) occurring in China (1-3). HCC is one of the most common forms of cancer in China and thus underscoring the urgent need for novel therapeutic treatment strategies for this deadly disease.
Citron kinase (CIT) is typed as an AGC (cAMP-dependent, cGMP-dependent and protein kinase C) protein kinase, and it is influenced by second messengers, including lipids such as PKC and cyclic AMP (4). The target proteins directly phosphorylated by CIT remain to be identified, although it is known to interact with proteins including KIF14 and TUBB3 (5,6). Known roles for CIT include a relationship with neurogenic cytokinesis, and this gene is known to be mutated in primary microcephaly, suggesting a key role for this gene in central nervous system development (7). A single nucleotide polymorphism in CIT has been found to be significantly related to schizophrenia, and the interaction between polymorphisms in CIT, NDEL1, and DISC1 is known to influence schizophrenia risk (8). CIK can also induce HIV-1 virion production by promoting Gag ubiquitination as well as improving viral release via the multivesicular bodies pathway (9). With respect to cancer, CIT appears to be related with the time to progression of ovarian cancer, and is also associated with therapeutic outcomes (10). CIT has been found to be overexpressed in human colon cancer tissues, wherein it may accelerate cancer cell growth via influencing the p53 signaling pathway (11). CIT protein is also overexpressed in breast cancer tissues, where it is associated with more aggressive forms of this disease (12). CIT depletion can mediate a failure of cytokinesis that may therefore valuable therapeutic efficacy as an anti-cancer strategy in cervical, breast, and colorectal cancer, and potentially in additional cancers (13).
Given the high incidence of HCC and the myriad roles of CIT, we were interested in exploring the role of CIT in HCC. We therefore investigated the expression of CIT in 235 HCC tissues and 96 non-tumorous liver tissues by immunohistochemistry staining to examine the correlations between CIT expression and clinicopathological parameters, as well as to compare this expression with overall patient survival rate. We found that CIT expression was associated with gender, tumor size, Edmondson Grade, Microvascular invasion, serum AFP level, and poor overall survival. We further reviewed available datasets in the Oncomine and UALCAN Expression Array databases to assess CIT mRNA expression. Unexpectedly, we found CIT mRNA expression to be higher in HCC tissues relative to normal liver tissue controls, leading us to speculate regarding the potential reasons for this discrepancy.
Methods
Patients and tissue samples
All the human tissues were acquired from HCC patients at Zhejiang Provincial People’s Hospital (Hangzhou, China). This research was approved by the Ethics Committee of Zhejiang Provincial People’s Hospital (Hangzhou, China). All the patients provided written informed consent.
A total of 235 paraffin-embedded HCC tissue samples and 96 non-tumorous liver tissue samples were acquired from April 2008 to September 2014. This patient cohort consisted 191 males and 44 females. Survival time was calculated based on the time between the date of surgery and the end of follow-up or date of death. All the tissues were used for a tissue microarray (TMA) analysis constructed by Shanghai Biochip Co., Ltd (Shanghai, China).
Immunohistochemical staining and evaluation
According to the manufacturer’s instructions, immunohistochemical staining was conducted using the Histostain-Plus IHC Kit (cat.no.856143; Invetrogen, USA). We heated 5 µm sections from the TMAs at 70 °C for 2 hours, after which they were de-paraffinized, rehydrated, and boiled in TE buffer for 3 min to retrieve antigen. Next, the sections were blocked with 3% H2O2 for 15 min to inhibit endogenous peroxidase activity, and they were then incubated with 10% goat non-immune serum for 20 min to decrease background non-specific staining. After that, samples were probed with a rabbit anti-CIT polyclonal antibody (1:400; lot. no. GR48652-1; ab110897; Abcam, Cambridge, UK) at 4 °C overnight, followed by incubation with a biotin-labeled secondary antibody for 20 min at room temperature, and then with HRP-conjugated streptavidin for 20 min at room temperature. A DAB Kit (ZSGB-BIO, Beijing, China) was additionally used to enhance color development. Finally, the sections were counterstained with hematoxylin, dehydrated, cleared, and mounted.
Two independent pathologists reviewed and scored all samples based on the strength of staining and on the percent of positively stained cells. A four-tiered scoring system was used as followed: for staining, 0 = negative, 1 = weak, 2 = moderate, and 3 = strong; for cell staining positivity: 0 for no cell stained, 1 for 1–25% of cells stained, 2 for 26–50% of cell stained, 3 for more than 50% of cells stained. As all sections tended to exhibit >50% staining in both HCC tissues and non-tumorous liver tissues, scores for the percent of positively stained cells were 3 for all samples. Scores for strength and percent of positively stained cells were multiplied together to yield an overall score. Scores <6 was indicative of low CIT expression, while scores ≥6 were indicative of high CIT expression.
Oncomine and UALCAN database analyses
A comprehensive analysis of extant datasets in the Oncomine Expression Array database (www.oncomine.org) was conducted to compare the mRNA expression of CIT between HCC and normal tissues, using the following search items: ‘CIT’, ‘mRNA’, ‘Cancer vs. Normal Analysis’ and ‘Hepatocellular Carcinoma’. We identified five relevant datasets including Chen Liver (14), Roessler Liver, Roessler Liver 2 (15), Mas Liver (16), and Wurmbach Liver (17). An additional analysis based on the online UALCAN database analysis (ualcan.path.uab.edu) was also conducted as above, unsing the search items: ‘Analysis’, ‘CIT’, ‘Liver Hepatocellular Carcinoma’.
Statistical analysis
Statistical analysis was fulfilled by using SPSS v13.0 (SPSS Inc., Chicago, IL). Chi-squared tests were used to assess the statistical significance of the relationship between CIT protein expression and clinicopathological parameters. We additionally used the Kaplan-Meier method to estimate survival curves, and the log-rank test was used for calculating differences between these curves. P<0.05 was the threshold of statistical significance.
Results
Expression of CIT in HCC and adjacent non-cancerous tissues
Immunostaining for CIT was evident in the cytoplasm and absent in the nuclei of both non-cancerous liver and HCC tissues. CIT was highly expressed in 94 of the 96 (97.91%) samples of adjacent non-cancerous liver tissues (Table 1). CIT expression was clearly decreased in HCC tissues, with low expression of CIT in 174 of the 235 (74.04%) HCC tissues (Figures 1,2). These decrease in CIT expression was significant (P<0.001).
Table 1
| Samples | CIT expression | P | |
|---|---|---|---|
| Low | High | ||
| Non-cancerous liver tissues | 2 | 94 | <0.001 |
| HCC tissues | 61 | 174 | |
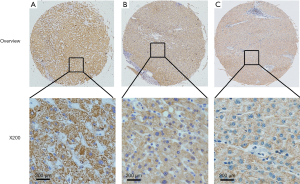
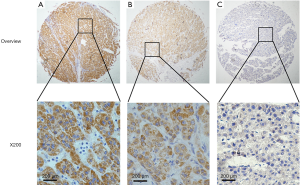
Relationship between CIT expression and clinicopathologic parameters
The relation between the expression of CIT and clinical variables was explored, and the results are shown in Table 2. CIT expression was significantly associated with gender, tumor size, Edmondson Grade, microvascular invasion, and AFP. The expression of CIT was significantly decreased in tumor of females, large tumors, tumors with a high Edmondson Grade, tumors with evident microvascular invasion or those with a high AFP level. There was no significant relationship between CIT expression and other assessed clinicopathologic parameters. These parameters and lower CIT expression are thus associated with poorer patient outcomes.
Table 2
| Clinical parameters | All cases | CIT | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.240 | |||
| <55 | 93 | 28 | 65 | |
| ≥55 | 142 | 33 | 109 | |
| Gender | 0.033 | |||
| Male | 191 | 44 | 147 | |
| Female | 44 | 17 | 27 | |
| Size | 0.017 | |||
| <5 | 111 | 21 | 90 | |
| ≥5 | 119 | 39 | 80 | |
| Tumour number | 0.726 | |||
| Single | 193 | 51 | 142 | |
| Multiple | 42 | 10 | 32 | |
| Edmondson grade | 0.011 | |||
| I+II | 151 | 31 | 120 | |
| III | 84 | 30 | 54 | |
| Metastasis | 0.180 | |||
| M0 | 212 | 52 | 160 | |
| M1 | 18 | 7 | 11 | |
| Microvascular invasion | 0.015 | |||
| Absence | 85 | 17 | 68 | |
| Presence | 99 | 36 | 63 | |
| HBs antigen | 0.123 | |||
| Negative | 46 | 16 | 30 | |
| Positive | 186 | 44 | 142 | |
| AFP | 0.003 | |||
| <50 | 96 | 10 | 86 | |
| ≥50 | 82 | 23 | 59 | |
| Status | <0.001 | |||
| Alive | 75 | 13 | 62 | |
| Dead | 54 | 26 | 28 | |
Survival analysis
The 5-year cumulative survival rate of patients with low CIT expression was 33.3%, while that of patients with high CIT expression was 68.9% based on a Kaplan-Meier survival analysis. Patients with low CIT expression had a mean survival time of 26.706±7.563 months, which was significantly shorter than that of patients in the high CIT expression group (45.860±4.449, P<0.001). These data indicated that low expression of CIT is related with poor overall survival (Figure 3).
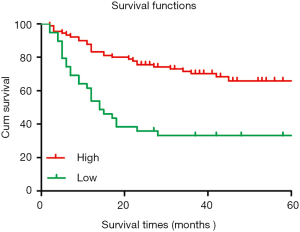
Analysis of CIT expression according to Oncomine and UALCAN databases
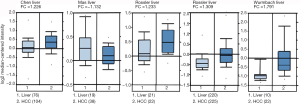
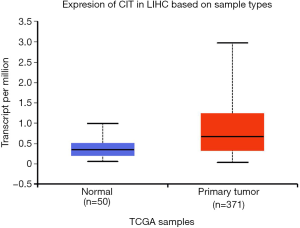
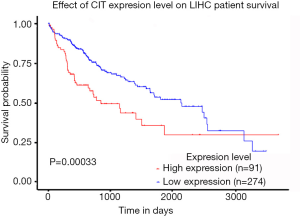
We next explored available datasets in the Oncomine database to compare CIT mRNA expression in HCC with normal tissues. What we found is that CIT mRNA expression was higher in HCC tissues relative to normal controls (Figure 4, all P<0.05, except P=0.980 between HCC and Mas Liver). Similarly, we found CIT mRNA expression to be higher in HCC tissues compared with normal tissues in the UALCAN Expression Array database (Figure 5, P<0.05). Survival analyses of these HCC patients showed that the patients with high CIT mRNA expression had significantly shorter survival times as compared with patients in the low CIT expression group based on the UALCAN database. These data thus demonstrate that high CIT mRNA expression was related to with poorer overall survival (Figure 6, P<0.05). These data highlight a discrepancy between these mRNA data and our protein level data, leading us to speculate that low CIT protein levels may mediate a feedback loop that results in a compensatory over-expression of the CIT mRNA.
Discussion
HCC is among the most common tumors in the world, and there is thus an urgent need to better understand and develop novel treatments for this disease. CIT is a multi-functional protein that is most highly expressed in fetal liver, with levels gradually decreasing after birth. Src and CIT are also downstream effectors of the Eph-induced signal transduction cascade, and Eph kinase activity control abscission (18). Indeed, CIT is known to be tightly linked with cytokinesis, a process which it controls using its coiled-coil domain, mediating the transition from constriction to abscission (19). Low level of TUBB3 in mitotic cells can be detrimental to their cytokinesis, and CIT can control TUBB3 phosphorylation to stabilize mid-body microtubules and cytokinesis (6). Two-pore channel 1 and p27Kip1 (p27) also interact with CIT to regulate cytokinesis (20,21). We had found no evidence for CIT to play a role in other mitotic events besides cytokinesis, until a recent research reported that it is required for the orientation of the mitotic spindle during metaphase (22). Aurora B, ASPM and CIT control mid-body architecture and spindle positioning during cytokinesis, and are necessary to maintain proliferating cell division (22,23). Specific tyrosine residues in CIT are known to be phosphorylated, and it has been demonstrated that such tyrosine phosphorylation of CIT impairs cytokinesis (19). CIT is an important abscission regulator, using active RhoA and anillin to promote mid-body stability (24). In our research, we found CIT to be expressed at lower levels in HCC tissues and at higher levels in normal tissues. This result suggests that CIT may function as a tumor suppressor in HCC, and may thus be an ideal therapeutic target in this disease. We thus conclude that lower CIT levels are associated with poorer patient outcomes in HCC.
Interestingly, while we observed a decrease in CIT levels in HCC samples at the protein level, we observed the opposite finding when assessing CIT mRNA levels in normal and HCC tissues using the Oncomine and UALCAN databases. Indeed, these results suggested that higher CIT expression was associated with poorer HCC prognosis, and a published article has also detected elevated CIT mRNA levels in HCC (25). These results are thus at odds with our protein level finding, which may be due to a difference in the readout being examined. Indeed, it may be that the decrease CIT protein levels which we observed in the present study initiated a feedback loop, resulting in increasing CIT mRNA expression in order to compensate for the decrease protein levels within cells, although further research will be needed to verify this finding.
In conclusion, we found that low CIT protein expression is associated with poorer outcomes in HCC patients. Low CIT expression was associated with gender, tumor size, Edmondson Grade, microvascular invasion, serum AFP level, and poor overall survival. Differences in the specific readout being examined may explain the discrepancies in the data between the present study and published databases.
Acknowledgments
Funding: This work was supported by the grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.58). All authors reports grants from Zhejiang Province Bureau of Health, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was allowed by Review Board of Hospital Ethics Committee, and the informed consent from every patient was obtained before we collected the data. Written informed consent was obtained from the patients to publish this paper. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Are C, Meyer B, Stack A, et al. Global trends in the burden of liver cancer. J Surg Oncol 2017;115:591-602. [Crossref] [PubMed]
- Wong MCS, Huang JLW, George J, et al. The changing epidemiology of liver diseases in the Asia–Pacific region. Nat Rev Gastro Hepat 2019;16:57-73. [Crossref] [PubMed]
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 2010;11:9-22. [Crossref] [PubMed]
- Li H, Bielas SL, Zaki MS, et al. Biallelic Mutations in Citron Kinase Link Mitotic Cytokinesis to Human Primary Microcephaly. Am J Hum Genet 2016;99:501-10. [Crossref] [PubMed]
- Sgro F, Bianchi FT, Falcone M, et al. Tissue-specific control of midbody microtubule stability by Citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ 2016;23:801-13. [Crossref] [PubMed]
- Basit S, Al-Harbi KM, Alhijji SA, et al. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum genet 2016;135:1199-207. [Crossref] [PubMed]
- Nicodemus KK, Callicott JH, Higier RG, et al. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet 2010;127:441-52. [Crossref] [PubMed]
- Ding J, Zhao J, Sun L, et al. Citron kinase enhances ubiquitination of HIV-1 Gag protein and intracellular HIV-1 budding. Arch Virol 2016;161:2441-8. [Crossref] [PubMed]
- Ehrlichova M, Mohelnikova-Duchonova B, Hrdy J, et al. The association of taxane resistance genes with the clinical course of ovarian carcinoma. Genomics 2013;102:96-101. [Crossref] [PubMed]
- Wu Z, Zhu X, Xu W, et al. Up-regulation of CIT promotes the growth of colon cancer cells. Oncotarget 2017;8:71954-64. [PubMed]
- Meng D, Yu Q, Feng L, et al. Citron kinase (CIT-K) promotes aggressiveness and tumorigenesis of breast cancer cells in vitro and in vivo: preliminary study of the underlying mechanism. Clin Transl Oncol 2019;21:910-23. [Crossref] [PubMed]
- McKenzie C, D’Avino PP. Investigating cytokinesis failure as a strategy in cancer therapy. Oncotarget 2016;7:87323-41. [Crossref] [PubMed]
- Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929-39. [Crossref] [PubMed]
- Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202-12. [Crossref] [PubMed]
- Mas VR, Maluf DG, Archer KJ, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med 2009;15:85-94. [Crossref] [PubMed]
- Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938-47. [Crossref] [PubMed]
- Jungas T, Perchey RT, Fawal M, et al. Eph-mediated tyrosine phosphorylation of citron kinase controls abscission. J Cell Biol 2016;214:555-69. [Crossref] [PubMed]
- Watanabe S, De Zan T, Ishizaki T, et al. Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J Cell Sci 2013;126:1773-84. [Crossref] [PubMed]
- Serres MP, Kossatz U, Chi Y, et al. Roberts JM, Malek NP and Besson A: p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J Clin Invest 2012;122:844-58. [Crossref] [PubMed]
- Horton JS, Wakano CT, Speck M, et al. Two-pore channel 1 interacts with citron kinase, regulating completion of cytokinesis. Channels (Austin) 2015;9:21-9. [Crossref] [PubMed]
- Gai M, Bianchi FT, Vagnoni C, et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep 2016;17:1396-409. [Crossref] [PubMed]
- McKenzie C, Bassi ZI, Debski J, et al. Cross-regulation between Aurora B and Citron kinase controls midbody architecture in cytokinesis. Open Biol 2016;6:160019. [Crossref] [PubMed]
- Gai M, Camera P, Dema A, et al. Citron kinase controls abscission through RhoA and anillin. Mol Biol Cell 2011;22:3768-78. [Crossref] [PubMed]
- Fu Y, Huang J, Wang KS. RNA interference targeting CITRON can significantly inhibit the proliferation of hepatocellular carcinoma cells. Mol Biol Rep 2011;38:693-702. [Crossref] [PubMed]


