
Spred-3 mutation and Ras/Raf/MAPK activation confer acquired resistance to EGFR tyrosine kinase inhibitor in an EGFR mutated NSCLC cell line
Introduction
Lung cancer is the most common malignancy and the primary cause of cancer-related deaths worldwide (1). This malignancy is mainly classified into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) (1), and NSCLC consists of 80% to 85% of all lung cancer patients (2). It is estimated that 15% of the NSCLC patients harbor activating mutations in epidermal growth factor receptor (EGFR) (3), and they are susceptible to EGFR tyrosine kinase inhibitors (TKIs), a new class of targeted therapeutic agents against lung cancer (4). Currently, identification of EGFR mutation is a standard procedure during the treatment of advanced NSCLC patients (5). Although survival benefits are achieved from EGFR-TKIs treatment in advanced NSCLC patients (6-8), acquired resistance to EGFR-TKIs in relapsed patents often occurs (9-11). T790M secondary mutation in EGFR and MET gene amplification have been shown to correlate with acquired resistance to EGFR-TKIs (12-14). Dysregulation of microRNA (miRNA) is also involved in acquired EGFR-TKI resistance (15-17). Overexpression of miR-21 has been linked to EGFR-TKI resistance in NSCLC because miR-21 can directly down-regulate PTEN and PDCD4 resulting in the activation of the PI3K/Akt pathway (18). Down-regulation of miR-205 has also been reported to induce erlotinib resistance in EGFR-mutated NSCLC (19). The miR-200c/LIN28B axis has also been shown to play an important role in NSCLC cells with acquired resistance to EGFR-TKI (20). Up-regulation of miR-21, AmiR-27a, and miR-218 may contribute to the primary resistance to EGFR-TKI in refractory advanced NSCLC patients with EGFR exon 19 deletion mutations (21). Our previous study has shown that down-regulation of miR-214 may reverse acquired resistance to EGFR-TKI in NSCLC through de-repressing LHX6 expression (22). Nonetheless, elucidation of the mechanisms underlying acquired EGFR-TKIs resistance remains incomplete.
Recently, whole-exome sequencing has been widely applied in cancer diagnosis and treatment (23-25). By using whole-exome sequencing, CSMD3 has been identified as the second most frequently mutated gene, next to TP53, in lung cancer (26). Single-nucleotide polymorphisms (SNPs) in the IRF1 and CYP181 genes and a frameshift deletion in the ALPK2 gene may be primarily responsible for the lack of efficacy in TKI treatment as well as relapse of chronic myeloid leukemia patients (27). Thus, we proposed that whole-exome sequencing would be instrumental in the identification of critical mutations that may shed light on novel mechanisms underlying the acquired resistance to EGFR-TKIs in EGFR-mutant NSCLC patients.
In this study, we identified gene mutations by whole-exome sequencing in NSCLC HCC827 cells that are sensitive to erlotinib, an EGFR-TKI, and erlotinib-resistant HCC827 (HCC827/ER) cells (22), and explored the molecular mechanisms underlying the acquired resistance to erlotinib in these NSCLC cells.
Methods
Cell culture and induction of erlotinib resistance in NSCLC cell lines
NSCLC HCC827 cell line harboring EGFR exon 19 deletion mutation was purchased from American Type Culture Collection (Manassas, VA, USA) and incubated in RPMI 1640 medium (Gibco; Garlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Garlsbad, CA, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin, at 37 °C, in a humidified atmosphere containing 5% CO2. HCC827 cells were subjected to erlotinib (Selleckchem; Houston, TX, USA) treatment by dose escalation for 6 months to generate erlotinib-resistant HCC827 cells (HCC827/ER cells). Drug resistance was measured by exposing the cells to various concentrations of erlotinib for 72 h in culture and cell viability was assessed by using the MTS assay to obtain the half maximal inhibitory concentration (IC50 value).
Whole-exome sequencing
Genomic DNA was extracted from the HCC827/ER cells and sheared with double-stranded DNA (dsDNA) fragmentase (New England Biolabs; Ispswich, MA, USA). Size selection (50 to 250 bp) was performed with Agencourt AMPure XP beads (Beckman Coulter, Inc.; Brea, CA, USA), and DNA fragments were used for library construction using the KAPA library preparation kit (Kapa Biosystems, Inc.; Wilmington, MA, USA) following the manufacturer’s instructions. All clean-up reactions were done with the Agencourt AMPure XP beads. Following fragmentation, the end repair and 3'-end dA-tailing was performed. Specifically, we used T-tailed adapters and enzymatically added a 3'-dA overhang on the fragmented DNA samples. Ligation was performed at 20 °C for 15 min. Single-step size selection was performed by adding 50 µL of PEG/NaCl SPRI Solution buffer (1×) to enrich ligated DNA fragments. The ligated fragments were then amplified by using 1× KAPA HiFi Hot Start Ready Mix and pre-capture ligation-mediated PCR (LM-PCR) Oligos in a 50 µL of reaction system and 7 to 8 PCR cycles, depending on input DNA mass. Library purity and concentration was quantified with a Qubit® 2.0 spectrophotometer (Life Technologies; Foster City, CA, USA). Fragment length was determined on a 2100 Bioanalyzer with the DNA 1000 Kit (Agilent Technologies, Inc.; Wilmington, DE, USA).
Hybrid selection was performed with a SeqCap EZ Exome Enrichment Kit version 3.0 (Roche NimbleGen; Madison, WI, USA). The hybridization of the amplified sample libraries and the SeqCap EZ Library was used at 47 °C for 16 to 20 h according to the manufacturer’s protocol. Following hybrid selection, the captured DNA fragments were amplified with 12 to 14 cycles of PCR using 1× KAPA HiFi Hot Start Ready Mix and post-capture LM-PCR Oligos in two separate 50 µL of reactions. The reactions were then pooled and purified with Agencourt AMPure XP beads. Library purity and concentration was assessed with a Qubit® 2.0 spectrophotometer. Fragment length was determined on a 2100 Bioanalyzer using the DNA 1000 Kit.
Multiplexed libraries were denatured by Tris-HCl and diluted by 0.2 N NaOH according to the manufacturer’s protocol (Illumina, Inc.; San Diego, CA, USA). Then, the libraries were sequenced using 150-bp paired-end runs on an Illumina Nextseq 500 sequencing system (Illumina, Inc.; San Diego, CA, USA). Sequences were aligned for variant calling and annotation with the human genome reference sequence (hg19 build) using BWA aligner. Downstream processing was performed with the genome analysis toolkit (GATK), SAMtools, and Picard Tools (http://picard.sourceforge.net/). A GATK UnifiedGenotyper and a GATK IndelGenotyperV2 were used to identify substitution and indel variant calls, respectively. Calls with a read coverage of ≤2 × and a Phred-scaled SNP quality of ≤20 were filtered out. All variants were annotated with the GATK Genomic Annotator (Broad Institute; Cambridge, MA, USA).
Quantitative real-time PCR (qPCR) assay
HCC827 and HCC827/ER cells were seed onto 6-well plates (NalgeNunc International Corporation; Rochester, NY, USA) at a density of 5×105 cells per well. Log-phase cells were digested in 0.25% Trypsin-EDTA (Gibco; Rockville, MD, USA), and then washed twice in PBS. Total RNA was extracted using a RNeasy Plus Mini kit (Qiagen, Inc.; Valencia, CA, USA), and RNA concentration was quantified with the Nanodrop® ND-1000 Spectrophotometer (Thermo Fisher Scientific; Waltham, MA, USA). Approximately 1 µg of total RNA sample was transcribed reversely into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific; Waltham, MA, USA) and Spred-3 expression was determined with the Lightcycler 480 SYBR Green I Master (Roche Applied Science; Indianapolis, IN, USA) under the following conditions: at 95 °C for 15 min, followed by 40 cycles of at 95 °C for 15 s, at 55 °C for 30 s and at 72 °C for 1 s, while β-actin served as an internal control (primers are shown in Table 1). Relative quantity of Spred-3 mRNA expression was calculated by using the 2−ΔΔCt method. All measurements were repeated in triplicate.
Table 1
| Primer | Sequence |
|---|---|
| Spred-3-F | 5'-TGGACTGACGTTTCAGAGC-3' |
| Spred-3-R | 5'-CCTGAAGCTGACTCCATCGT-3' |
| β-actin-F | 5'-CATCCGCAAAGACCTGTACG-3' |
| β-actin-R | 5'-CATCCGCAAAGACCTGTACG-3' |
Spred-3 knockout and over-expression in cells
Two pairs of oligonucleotides were designed based on the sequence of exon 1 of the Spred-3 gene using the online CRISPR software (http://crispr.mit.edu/), while AAV served as a control (oligonucleotide sequences are described in Table 2). The oligonucleotide pairs were ligated to the vector lentiCRISPRv2, and co-transfected with the packaging plus envelope plasmid PsPAX2 and the packaging plasmid pCMV-VSVG into 293T cells to generate lentiCRISPRv2-Spred-3-sg1, lentiCRISPR-Spred-3-sg2 and lentiCRISPR-AAV-sglentiviruses. HCC827 cells were seed onto 6-well plates at a density of 5×105 cells per well for 24 h. Then, log-phase cells were infected with lentiCRISPRv2-Spred-3-sg1, lentiCRISPR-Spred-3-sg2 and lentiCRISPR-AAV-sg viruses in RPMI 1640 medium containing 6 µg/mL polybrene. After 72 h infection, HCC827 cells were screened with 2 µg/mL puromycin for 2 weeks to generate stably transfected HCC827/AAV-sg, HCC827/Spred-3-sg1 and HCC827/Spred-3-sg2 cells.
Table 2
| Oligonucleotide | Sequence |
|---|---|
| Spred-3-sg1-Oligo1 | 5'-CACCGTGAGCGTGTGTCGGGTCCGA-3' |
| Spred-3-sg1-Oligo2 | 5'-AAACTCGGACCCGACACACGCTCAC-3' |
| Spred-3-sg2-Oligo1 | 5'-CACCGTCATCCACGGGGAACGCCTC-3' |
| Spred-3-sg2-Oligo2 | 5'-AAACGAGGCGTTCCCCGTGGATGAC-3' |
| AAV-Oligo1 | 5'-CACCGGGGCCACTAGGGACAGGAT-3' |
| AAV-Oligo2 | 5'-AAACATCCTGTCCCTAGTGGCCCC-3' |
The CDS sequence of the Spred-3 gene was captured from the UCSC genome browser (http://genome.ucsc.edu/) to construct the vector pLVX-IRES-ZsGreen1, which was then co-transfected with the packaging plus envelope plasmid PsPAX2 and the packaging plasmid pCMV-VSVG into 293T cells to generate pLV-Spred-3 and pLV lentiviruses. HCC827/ER cells were seed onto 6-well plates at a density of 5×105 cells per well for 24 h. Then, log-phase cells were infected with pLV-Spred-3 and pLV viruses in RPMI 1640 medium containing 6 µg/mL polybrene. After 72 h infection, HCC827/ER cells were screened with 2 µg/mL puromycin for 7 days to generate stably transfected HCC827/ER/pLV-Spred-3 and HCC827/ER/pLV cells.
Western blotting assay
HCC827 and HCC827/ER cells were seeded onto 6-well plates at a density of 5×105 cells per well for 24 h. Then, the medium was discarded, and log-phase cells were incubated in serum-free medium containing EGF (Invitrogen; Carlsbad, CA, USA) at concentrations of 0, 50, 100 and 200 ng/mL for 6 h. HCC827/AAV-sg and HCC827/Spred-3-sg were seeded onto 6-well plates at a density of 5×105 cells per well for 24 h. Then, the medium was discarded, and log-phase HCC827/AAV-sg cells were incubated in serum-free medium, while log-phase HCC827/Spred-3-sg cells were incubated in serum-free medium containing 4 nM SCH772984 (Selleckchem; Houston, TX, USA), an ERK1/2 inhibitor for 30 min. HCC827, HCC827/AAV-sg, HCC827/Spred-3-sg1, HCC827/Spred-3-sg2, HCC827/ER, HCC827/ER/pLV and HCC827/ER/pLV-Spred-3 cells were harvested, digested in 0.25% Trypsin-EDTA, washed twice in PBS, and completely lysed in cell lysis buffer (Cell Signaling Technology; Beverly, MA, USA) on ice. The solution was centrifuged at 12,000 g at 4 °C for 10 min and the supernatant was collected. The concentration of total protein was quantified with the BCA Protein Assay Kit (Thermo Fisher Scientific; Waltham, MA, USA) and 30 µg of the total protein was separated with SDS-PAGE. Subsequently, the blots were transferred to nitrocellulose (NC) membranes and blocked in 3% bovine serum albumin (BSA) at room temperature for 1 h. Then, the blots were incubated in TBST (20 mM Tris-HCl, 150 mM NaCl and 0.05% Tween-20; pH 7.4) containing rabbit anti-Spred-3 polyclonal antibody (1:1,000 dilution; Sigma-Aldrich, St Louis, MI, USA), mouse anti-phospho-p44/42 MAPK (ERK1/2) monoclonal antibody (1:1,000 dilution; Cell Signaling Technology; Beverly, MA, USA), rabbit anti-p44/42 MAPK (ERK1/2) polyclonal antibody (1:1,000 dilution; Cell Signaling Technology; Beverly, MA, USA), rabbit anti-AKT polyclonal antibody (1:1,000 dilution; Cell Signaling Technology; Beverly, MA, USA) and mouse anti-phospho-AKT (Ser473) monoclonal antibody (1:1,000 dilution; Cell Signaling Technology; Beverly, MA, USA) at 4 °C overnight, while β-actin was used as a loading control. The blots were then washed three times in TBST, of 10 min each time, incubated in anti-rabbit/mouse HRP-conjugated IgG antibody (1:4,000 dilution; Cell Signaling Technology; Beverly, MA, USA) at room temperature for 2 h, and washed three times in TBST, of 10 min each time. The protein bands were visualized using an ECL Kit (Thermo Fisher Scientific; Waltham, MA, USA), and the expression level of each protein was normalized to that of β-action. All measurements were repeated in triplicate.
Transwell invasion assay
HCC827, HCC827/AAV-sg, HCC827/Spred-3-sg2, HCC827/ER, HCC827/ER/pLV and HCC827/ER/pLV-Spred-3 cells were digested in 0.25% Trypsin-EDTA (Gibco; Rockville, MD, USA), and adjusted to a density of 5×103 cells/mL in serum-free RPMI 1640 medium. Approximately 100 µL of cells were transferred to the upper Transwell chamber (Millipore; Bedford, CA, USA), and the lower chamber was coated with 1 mg/ml fibronetin. The Transwell chambers were inserted into 24-well plates (Corning, Inc.; Corning, NY, USA), and the lower chamber was immersed in the RPMI 1640 medium containing 20% FBS. Following incubation at 37 °C for 48 h, the Transwell chambers were removed from the 24-well plate and washed twice in PBS. Then, cells were gently removed from the upper chamber using cotton swabs, fixed in methanol for 15 min, stained with 0.1% crystal violet and dried. Five fields of vision were randomly selected, and the number of cells in each field was counted under a microscope at a magnification of ×200. All experiments were repeated in triplicate.
MTS assay
HCC827, HCC827/AAV-sg, HCC827/Spred-3-sg2, HCC827/ER, HCC827/ER/pLV and HCC827/ER/pLV-Spred-3 cells were seeded onto 96-well plates at a density of 5×103 cells per well and cultured for 24 h. Then, log-phase cells were incubated in 100 µL RPMI 1640 medium containing erlotinib at concentrations of 0.1, 0.2, 0.4, 0.8 and 1.6 µM. After 72 h culture, the medium was discarded, and each well was added with 100 µl of the McCoy’s 5A medium and 20 µL of the MTS solution (Promega; Madison, WI, USA). Following incubation at 37 °C for 2 h, the optical density (OD) was measured at 490 nm with the Model 680 microplate reader (Bio-Rad Laboratories; Hercules, CA, USA). HCC827/AAV-sg and HCC827/Spred-3-sg cells were seeded onto 96-well plates at a density of 5×103 cells per well and cultured for 24 h. Then, log-phase HCC827/AAV-sg and HCC827/Spred-3-sg cells were incubated in 100 µL RPMI 1640 medium containing erlotinib at concentrations of 0.1, 0.2, 0.4, 0.8 and 1.6 µM, and HCC827/Spred-3-sg cells were incubated in 100 µL RPMI 1640 medium containing 4 nM ERK1/2 inhibitor SCH772984 and erlotinib at concentrations of 0.1, 0.2, 0.4, 0.8 and 1.6 µM. After 72 h culture, the medium was discarded, and each well was added with 100 µL of the McCoy’s 5A medium and 20 µL of the MTS solution. Following incubation at 37 °C for 2 h, the OD was measured at 490 nm with the Model 680 microplate reader. The half maximal inhibitory concentration (IC50) of erlotinib against the NSCLC cells was calculated with the statistical software SPSS version 20.0 (SPSS, Inc.; Chicago, IL, USA). All experiments were repeated in triplicate.
Data analysis
All data were expressed as mean ± standard deviation (SD), and all statistical analyses were done using the software GraphPad Prism version 6.0. Differences of means between groups were tested for statistical significance with Student t test, and a P value <0.05 was considered statistically significant.
Results
A frameshift and loss-of-function mutation at exon 1 of the Spred-3 gene was identified in HCC827 cells with acquired erlotinib resistance
Erlotinib resistant (HCC827/ER) cells were obtained by culturing the HCC827 cell line with dose escalation of the drug. IC50 was then measured by exposing the cells to erlotinib at concentrations of 0, 0.1, 0.2, 0.4, 0.8, and 1.6 µM. Using the MTS assay, the IC50 of HCC827/ER cells was found to be nearly 7-fold higher than the IC50 of HCC827 (1.96 vs. 0.26 µM). To identify genomic mutations that might contribute to the drug resistance, whole-exome sequencing was conducted in the HCC827/ER cells. Among twenty-some mutations detected, a deletion mutation (c.120delG) at exon 1 of the Spred-3 gene, which would result in a frameshift mutation and premature translation termination (p.E40fs), was identified with an extremely high frequency in HCC827/ER cells (Figure 1A, Table 3). This high frequency suggested the Spred-3 mutation occurred very early during the drug resistance selection process. Thus, we decided to characterize this Spred-3 mutation and to investigate whether this mutation contributed to drug resistance.
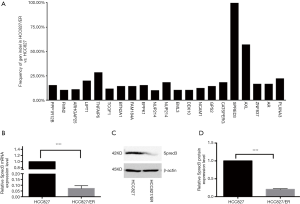
Table 3
| Gene | Transcript accession number | Exon | Base changes | Amino acid change | Mutation rate (%) |
|---|---|---|---|---|---|
| PPP1R12 | NM_001167857 | Exon 10 | c.1496delT | p.V499fs | 15.38 |
| FMN2 | NM_001305424 | Exon 1 | c.157_159del | p.53_53del | 10.53 |
| ARHGAP5 | NM_001166276 | Exon 8 | c.1081delA | p.K361fs | 11.11 |
| LIPT1 | NM_145199 | Exon 2 | c.361delA | p.K121fs | 20 |
| TNFAIP6 | NM_007115 | Exon 6 | c.833delA | p.X278X | 28.57 |
| TCOF1 | NM_000356 | Exon 23 | c.3853_3855del | p.1285_1285del | 11.76 |
| BTN2A1 | NM_001197233 | Exon 7 | c.1255delT | p.F419fs | 14.29 |
| FAM184A | NM_001100411 | Exon 2 | c.53_54insGG | p.D18fs | 14.29 |
| EPPK1 | NM_031308 | Exon 2 | c.6044delG | p.G2015fs | 15.38 |
| NUP214 | NM_005085 | Exon 29 | c.4127delC | p.A1376fs | 10 |
| NUP214 | NM_005085 | Exon 29 | c.4780delA | p.T1594fs | 18.18 |
| EML3 | NM_001300793 | Exon 3 | c.302delC | p.P101fs | 10.53 |
| DDX10 | NM_004398 | Exon 17 | c.2340_2342del | p.780_781del | 10.26 |
| NCAM1 | NM_000615 | Exon 1 | c.5delT | p.L2fs | 12.50 |
| GPS2 | NM_004489 | Exon 10 | c.868delC | p.Q290fs | 14.29 |
| CATSPERG | NM_021185 | Exon 17 | c.1894_1895del | p.L632fs | 18.18 |
| SPRED/3 | NM_001042522 | Exon 1 | c.120delG | p.E40fs | 100 |
| AXL | NM_001278599 | Exon 4 | c.27delC | p.D9fs | 57% |
| ZNF837 | NM_138466 | Exon 3 | c.469delC | p.R157fs | 16.67 |
| AR | NM_000044 | Exon 1 | c.171_182del | p.57_61del | 16.67 |
| PLXNA3 | NM_017514 | Exon 2 | c.42dupG | p.V14fs | 22.22 |
First, we investigated the expression of Spred-3 in HCC827/ER cells carrying the c.120delG deletion/frameshift mutation. To measure the Spred-3 mRNA, primers targeting the coding sequence were designed. qPCR assay detected a much lower level of Spred-3 mRNA in HCC827/ER than the level in HCC827 cells (P<0.0001) (Figure 1B). The decrease of Spred-3 mRNA expression in the HCC827/ER cells was presumably due to non-sense mediated decay of mutant Spred-3 mRNA. We then used Western blotting analysis to determine the Spred-3 protein expression, and found a similar decrease in the amount of Spred-3 protein in HCC827/ER as compared to the amount in HCC827 (P<0.0001) (Figure 1C,D). Thus, the resistant cells carrying the single nucleotide deletion expressed little amount of Spred-3.
Spred-3 has been identified as a suppressor of the Ras/Raf/MAPK pathway (28), so a decrease in the Spred-3 expression might result in an abnormal, constitutive activation of the Ras/Raf/MAPK pathway in HCC827/ER cells. We investigated this hypothesis and presented the finding in a later section.
Knocking out Spred-3 in HCC827 cells increases the resistance to erlotinib and cell migration
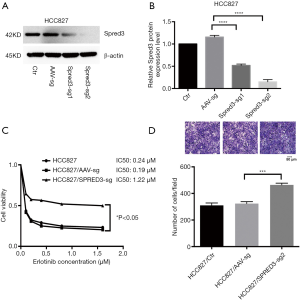
To investigate the contribution of Spred-3 to erlotinib resistance, we used the CRISPR/Cas9 system to inactivate the Spred-3 gene in HCC827 cells by creating insertion-deletion (indel) mutations in the coding sequence. HCC827 cells were transduced separately with two lentiviral vectors each carrying a different Spred-3 targeting sgRNA. Western blotting assay detected significantly lower amounts of Spred-3 in HCC827/Spred-3-sg1 and HCC827/Spred-3-sg2 cells than the amount of Spred-3 in the control HCC827/AAV-sg cells (P<0.0001) (Figure 2A,B). MTS assays showed 0.24, 0.19 and 1.22 µM erlotinib IC50 values for HCC827, HCC827/AAV-sg and HCC827/Spred-3-sg2 cells, respectively, and the viability in the presence of erlotinib of the HCC827/Spred-3-sg2 cells was found to be significantly higher than that of the HCC827/AAV-sg cells (P<0.05) (Figure 2C), indicating that loss of Spred-3 increased the resistance to erlotinib in HCC827 cells. In addition, Transwell migration assays showed a significant increase in the number of membrane-penetrating cells in HCC827/Spred-3-sg2 than the number in HCC827/AAV-sg (P<0.001) (Figure 2D), demonstrating that loss of Spred-3 increases the migration ability of HCC827 cells.
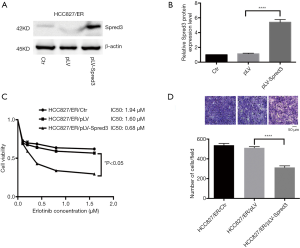
Ectopic expression of Spred-3 in HCC827/ER cells suppresses the resistance to erlotinib and cell migration
We ectopically expressed Spred-3 in HCC827/ER cells by lentiviral transduction with pLV-Spred-3. The amount of Spred-3 in HCC827/ER/pLV-Spred-3 cells detected by Western blotting assays was significantly higher than the amount detected in HCC827/ER/pLV cells (P<0.0001) (Figure 3A,B). MTS assays revealed 1.94, 1.6 and 0.68 µM erlotinib IC50 values for HCC827/ER, HCC827/ER/pLV and HCC827/ER/pLV-Spred-3 cells, respectively, and the viability in the presence of erlotinib of the HCC827/ER/pLV-Spred-3 cells was significantly lower than that of the HCC827/ER/pLV cells (P<0.05) (Figure 3C), indicating that re-expression of Spred-3 reduces the resistance to erlotinib in HCC827/ER cells. In addition, Transwell migration assays showed a significantly lower number of membrane-penetrating cells in HCC827/ER/pLV-Spred-3 than the number in HCC827/ER/pLV (P<0.0001) (Figure 3D), indicating that re-expression of Spred-3 suppresses HCC827/ER cell migration.
The sensitivity to erlotinib and cell migration is mediated via the Ras/Raf/MAPK pathway in HCC827 cells
As described above, whole-exome sequencing identified a c.120delG mutation at exon 1 of the Spred-3 gene in HCC827/ER cells, leading to a drastic decrease in the amount of full-length Spred-3 protein. Since Spred-3 is known to suppress the Ras/Raf/MAPK signaling pathway in response to epidermal growth factor (EGF), we decided to investigate the role of this signaling pathway in erlotinib resistance. We treated HCC827 and HCC827/ER cells in serum-free medium supplemented with dose escalation of EGF for 30 min. Western blotting assays detected increasing amounts of p-p44/42 and p-AKT in HCC827/ER cells upon elevating EGF concentrations, while the amounts of those phosphorylated proteins were essentially unchanged in HCC827 cells (Figure 4A,B,C,D). Also, it appeared that, prior to the EGF induction, the level of the phosphorylated p44/42 in HCC827/ER was already higher than the level in HCC827 (Figure 4A,B). Thus, these results suggested that the Ras/Raf/MAPK pathway may be de-repressed in HCC827/ER cells in the absence of functional Spred-3, and the pathway can be readily activated following EGF treatment. The findings support our hypothesis that HCC827 cells expressed Spred-3 that could suppress the activation of the Ras/Raf/MAPK pathway and HCC827/ER cells did not express functional Spred-3 so that the Ras/Raf/MAPK signaling could become activated upon EGF treatment.
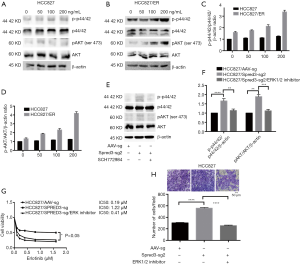
To further test our hypothesis, we performed Western blotting analysis and found higher p-p44/42 expression in HCC827/Spred-3-sg2 cells than in HCC827/AAV-sg cells (Figure 4E,F), indicating that the Ras/Raf/MAPK pathway could still be activated by EGF in HCC827 cells when Spred-3 was inactivated by a Spred-3 specific sgRNA. We also inhibited the Ras/Raf/MAPK pathway in HCC827/Spred-3-sg2 cells with an ERK1/2-specific inhibitor SCH772984, and Western blotting detected significant down-regulation of p-p44/42 expression when HCC827/Spred-3-sg2 cells were treated with SCH772984 than cells treated with the solvent (Figure 4E,F). MTS assays revealed 0.19, 1.22 and 0.41 µM erlotinib IC50 values for HCC827/AAV-sg, HCC827/Spred-3-sg2 and SCH772984-treated HCC827/Spred-3-sg2 cells, respectively, and the viability in the presence of erlotinib of the SCH772984-treated HCC827/Spred-3-sg2 cells appeared to be lower than that of the HCC827/Spred-3-sg2 cells (P=0.0974) (Figure 4G). Transwell migration assays showed a significant lower number of membrane-penetrating cells in SCH772984-treated HCC827/Spred-3-sg2 than the number in HCC827/Spred-3-sg2 (P<0.0001) (Figure 4H), indicating that inhibition of the Ras/Raf/MAPK pathway reduced the migration of HCC827/Spred-3-sg2 cells.
Discussion
EGFR-mutant NSCLC patients exhibit remarkable and persistent responses to EGFR-TKIs (29,30), such as gefitinib and erlotinib; however, disease progression may occur at a median response time of 12 months, because of the development of acquired drug resistance during the therapy (31,32). To date, gene mutations have been identified as the major mechanisms of EGFR-TKI resistance in NSCLC (3,4). Systematic genetic and histological analyses of tumor biopsies from 37 patients with EGFR-TKI resistant NSCLC carrying EGFR mutations using an SNaPshot assay, a multiplex platform that is used at Massachusetts General Hospital to genotype cancers at specific genetic loci across 13 genes, showed 49% patients with exon 20 EGFR mutation T790M, 5% with MET amplification, 5% with PIK3CA mutations, and 5% with β-catenin mutations (33). By this estimation, putative resistant mutations are not accounted for nearly one-third of the resistant patients.
In this study, whole-exome sequencing identified, for the first time, an indel mutation at exon 1 of the Spred-3 gene, which predicts a frameshift mutation at amino acid 40 and loss of functional protein, can result in resistance to an EGFR-TKI. Western blotting and qPCR assays detected little Spred-3 protein and mRNA expression in the drug resistant HCC827/ER cells. These findings suggest that a loss-of-function mutation in the Spred-3 gene can lead to acquired resistance to EGFR-TKIs in EFGR mutant cell lines and possibly NSCLC as well.
Spred-3, also known as Eve-3, is a peripheral membrane protein of 410 amino acids with a C-terminal Sprouty-like cysteine-rich domain (SRY) and an N-terminal Ena/Vasodilator-stimulated phosphoprotein (VASP) homology-1 (EVH-1) domain. Spred-3 is a member of a family of proteins that negatively regulates Ras/Raf/MAPK signaling, particularly during organogenesis (28). Spred-3 has been reported to be exclusively expressed in the brain (34), and the gene is located in chromosome 19q13.13 which is often deleted in glioblastoma; however, a somatic mutation study concluded that Spred-3 seems not to be a 19q13 glioblastoma gene (35). We have not retrieved Spred-3 mutation in NSCLC in the The Cancer Genome Atlas (TCGA) database (36), thus the role of Spred-3 in cancers remained unclear until this study.
In an unbiased screen, we identified in high frequency a Spred-3 deletion mutation in erlotinib resistant HCC827/ER cells. We then knocked out the Spred-3 gene in the drug-sensitive parental HCC827 cells using CRISPR/Cas9, and found the Spred-3 knockout cells had an increase in the resistance to erlotinib as well as cell migration. We also expressed Spred-3 ectopically in HCC827/ER cells, and found the Spred-3 expressing cells became less resistant to erlotinib and less efficient in cell migration. Our data demonstrate that Spred-3 is involved in the acquired resistance to EGFR-TKIs in NSCLC cells harboring EGFR mutations.
EGFR signaling plays a critical role during tumorigenesis through activating the downstream signaling pathway (37), in which Ras/Raf/MAPK and PI3K/AKT signaling are the two most important pathways (38). Gene mutations in the Ras/Raf/MAPK pathway have been shown to confer resistance to EGFR-TKIs in NSCLC cell lines (39). Here, we identified, for the first time, Spred-3 can be a contributor to the acquired EGFR-TKI resistance in NSCLC cells harboring EGFR mutations. Since Spred-3 is a suppressor of the MAPK/ERK1/2 pathway (28), it is consistent with this notion that we found abnormal activation and up-regulation of p-p44/42 and p-AKT expression in HCC827/ER cells with an EGF treatment. It appears that the inactivation of the Spred-3 protein resulted in the activation of the MAPK/ERK1/2 pathway and promoted the activation of the PI3K/AKT pathway in HCC827/ER cells, which is consistent with the existence of a crosstalk between the MAPK/ERK1/2 and PI3K/AKT pathways (40). Furthermore, ERK1/2 inhibitor treatment also led to down-regulation of the p-p44/42 and p-AKT expression in CRISPR-mediated Spred-3 knockout cells, indicating the suppression of the MAPK/ERK1/2 signaling pathway resulted in a remarkable reduction in EGFR-TKI resistance and cell migration. Taken together, these data demonstrated that Spred-3 inactivation could cause over-activation of the MAPK/ERK1/2 and PI3K/AKT pathways and increase EGFR-TKI resistance and cell migration in NSCLC cells.
The regulation involving EGFR, ERK, and AKT is summarized in Figure 5. EGFR-TKI works by binding to the EGFR and inhibiting ATP binding which resulted in suppressing EGFR autophosphorylation. This in turn inhibits EGFR binding to Grb and Sos, and inhibits Ras protein binding to the cell membrane, resulting in the inhibition of Ras/Raf/MEK/ERK and PI3K/AKT signaling pathways. As a result, TKI inhibits cell growth. Like Spred-1, Spred-3 binds to B-Raf/C-Raf on the cell membrane, and this prevents K-Ras from locating to the cytoplasmic membrane and suppresses the activation of ERK signaling (41). As such, Spred-3 suppresses the ERK signaling pathway. In HCC827/ER cells, EGFR-TKI may still bind EGFR, inhibits Ras protein binding to the cell membrane, and inhibits the Ras/Raf/MEK/ERK signaling pathway. However, the loss of functional Spred-3, due to the deletion mutation c.120delG at exon 1 of the Spred-3 gene, de-represses the B-Raf/C-Raf-mediated ERK signaling by allowing K-Ras to locate to the cytoplasmic membrane and to activate the ERK signaling pathway. Since there is a cross-talk between ERK signaling and PI3K/Akt pathway (42), the loss of Spred-3 will activate both ERK and PI3K/Akt to induce growth-related gene expression, thereby resulting in EGFR-TKI resistance.
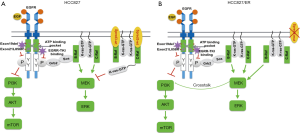
Conclusions
In summary, we identified an indel mutation at exon 1 of the Spred-3 gene in HCC827/ER cells generated by erlotinib treatment at dose escalation (22), and we detected a drastic decrease in the Spred-3 expression at both translational and transcriptional levels. Knockout of Spred-3 causes an increase in the resistance to erlotinib and in cell migration in HCC827 cells, while ectopic expression of Spred-3 reduces the resistance to erlotinib and cell migration in HCC827/ER cells. In addition, inhibition of the Ras/Raf/MAPK pathway increases the sensitivity to erlotinib and reduces the viability and migration in HCC827 cells. We concluded that Spred-3 mutation-induced activation of the Ras/Raf/MAPK pathway confers the acquired resistance to EGFR-TKIs in NSCLC cell lines harboring EGFR mutations. Further studies to investigate the prevalence of Spred-3 mutations in NSCLC patients with resistance to EGFR-TKI will be of great interest.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethical Review Committee of Fujian Provincial Cancer Hospital (approval no. FJZLYY2015-00179). All experimental procedures described in this study were conducted in accordance with national and local laws, regulations and guidelines.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Neal RD, Sun F, Emery JD, et al. Lung cancer. BMJ 2019;365:l1725. [Crossref] [PubMed]
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Castellanos E, Feld E, Horn L. Driven by mutations: The predictive value of mutation subtype in EGFR-mutated non-small cell lung cancer. J Thorac Oncol 2017;12:612-23. [Crossref] [PubMed]
- Doval DC, Desai CJ, Sahoo TP. Molecularly targeted therapies in non-small cell lung cancer: The evolving role of tyrosine kinase inhibitors. Indian J Cancer 2019;56:S23-30. [Crossref] [PubMed]
- Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol 2019;12:134. [Crossref] [PubMed]
- Inal C, Yilmaz E, Piperdi B, et al. Emerging treatment for advanced lung cancer with EGFR mutation. Expert Opin Emerg Drugs 2015;20:597-612. [Crossref] [PubMed]
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-59. [Crossref] [PubMed]
- Wang P, Zhang D, Li XM, et al. Continued EGFR-TKIs treatment promotes the survival of elderly patients with acquired resistance to EGFR-TKIs therapy. Eur Rev Med Pharmacol Sci 2016;20:2450-9. [PubMed]
- Martinez-Marti A, Navarro A, Felip E. Epidermal growth factor receptor first generation tyrosine-kinase inhibitors. Transl Lung Cancer Res 2019;8:S235-46. [Crossref] [PubMed]
- Andrews Wright NM, Goss GD. Third-generation epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small cell lung cancer. Transl Lung Cancer Res 2019;8:S247-64. [Crossref] [PubMed]
- Crinò L, Delmonte A. Molecular targeted therapies in non-small cell lung cancer: where we are. Transl Lung Cancer Res 2019;8:S222-3. [Crossref] [PubMed]
- Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [Crossref] [PubMed]
- Denis MG, Vallée A, Théoleyre S. EGFR T790M resistance mutation in non-small-cell lung carcinoma. Clin Chim Acta 2015;444:81-5. [Crossref] [PubMed]
- Wu YL, Soo RA, Locatelli G, et al. Does c-Met remain a rational target for therapy in patients with EGFR TKI-resistant non-small cell lung cancer? Cancer Treat Rev 2017;61:70-81. [Crossref] [PubMed]
- Zhang K, Yuan Q. Current mechanism of acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and updated therapy strategies in human nonsmall cell lung cancer. J Cancer Res Ther 2016;12:C131-7. [Crossref] [PubMed]
- Ahsan A. Mechanisms of resistance to EGFR tyrosine kinase inhibitors and therapeutic approaches: an update. Adv Exp Med Biol 2016;893:137-53. [Crossref] [PubMed]
- Ricciuti B, Mecca C, Cenci M, et al. miRNAs and resistance to EGFR-TKIs in EGFR-mutant non-small cell lung cancer: beyond 'traditional mechanisms' of resistance. Ecancermedicalscience. 2015;9:569. [Crossref] [PubMed]
- Li B, Ren S, Li X, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014;83:146-53. [Crossref] [PubMed]
- Park KS, Raffeld M, Moon YW, et al. CRIPTO1 expression in EGFR-mutant NSCLC elicits intrinsic EGFR-inhibitor resistance. J Clin Invest 2014;124:3003-15. [Crossref] [PubMed]
- Sato H, Shien K, Tomida S, et al. Targeting the miR-200c/LIN28B axis in acquired EGFR-TKI resistance non-small cell lung cancer cells harboring EMT features. Sci Rep 2017;7:40847. [Crossref] [PubMed]
- Wang S, Su X, Bai H, et al. Identification of plasma microRNA profiles for primary resistance to EGFR-TKIs in advanced non-small cell lung cancer (NSCLC) patients with EGFR activating mutation. J Hematol Oncol 2015;8:127. [Crossref] [PubMed]
- Liao J, Lin J, Lin D, et al. Down-regulation of miR-214 reverses erlotinib resistance in non-small-cell lung cancer through up-regulating LHX6 expression. Sci Rep 2017;7:781. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U. etal. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Beltran H, Eng K, Mosquera JM, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 2015;1:466-74. [Crossref] [PubMed]
- Li X, Yang M, Zhang QZ, et al. Whole exome sequencing in the accurate diagnosis of bilateral breast cancer: A case study. J Breast Cancer 2019;22:131-40. [Crossref] [PubMed]
- Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012;33:1270-6. [Crossref] [PubMed]
- Abedin S, Platanias LC. Whole-exome sequencing for relapse prediction in patients discontinuing TKI treatment in chronic myeloid leukemia. Leuk Lymphoma 2016;57:1503-4. [Crossref] [PubMed]
- King JA, Corcoran NM, D'Abaco GM, et al. Eve-3: a liver enriched suppressor of Ras/MAPK signaling. J Hepatol 2006;44:758-67. [Crossref] [PubMed]
- Rawluk J, Waller CF. Gefitinib. Recent Results Cancer Res 2018;211:235-46. [Crossref] [PubMed]
- Steins M, Thomas M, Geißler M. Erlotinib. Recent Results Cancer Res 2018;211:1-17. [Crossref] [PubMed]
- Lim SM, Syn NL, Cho BC, et al. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Treat Rev 2018;65:1-10. [Crossref] [PubMed]
- Fogli S, Polini B, Del Re M, et al. EGFR-TKIs in non-small-cell lung cancer: Focus on clinical pharmacology and mechanisms of resistance. Pharmacogenomics 2018;19:727-40. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Kato R, Nonami A, Taketomi T, et al. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem Biophys Res Commun 2003;302:767-72. [Crossref] [PubMed]
- Mizoguchi M, Nutt CL, Louis DN. Mutation analysis of CBL-C and SPRED-3 on 19q in human glioblastoma. Neurogenetics 2004;5:81-2. [Crossref] [PubMed]
- Wang Z, Jensen MA, Zenklusen JC. A practical guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41. [Crossref] [PubMed]
- Horne SD, Pollick SA, Heng HH. Evolutionary mechanism unifies the hallmarks of cancer. Int J Cancer 2015;136:2012-21. [Crossref] [PubMed]
- Seshacharyulu P, Ponnusamy MP, Haridas D, et al. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin Ther Targets 2012;16:15-31. [Crossref] [PubMed]
- Huang L, Fu LW. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B 2015;5:390-401. [Crossref] [PubMed]
- Zhou J, Du T, Li B, et al. Crosstalk between MAPK/ERK and PI3K/AKT signal pathways during brain ischemia/reperfusion. ASN Neuro 2015;7:1759091415602463. [Crossref] [PubMed]
- Siljamäki E, Abankwa D. SPRED1 interferes with K-ras but not H-ras membrane anchorage and signaling. Mol Cell Biol 2016;36:2612-25. [Crossref] [PubMed]
- De Luca A, Maiello MR, D'Alessio A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16:S17-27. [Crossref] [PubMed]


