
COPB2 promotes metastasis and inhibits apoptosis of lung adenocarcinoma cells through functioning as a target of miR-216a-3p
Introduction
According to the statistics released by International Agency for Research on Cancer in 2018, lung cancer has the highest global incidence (11.6%) and mortality rate (18.4%) compared with other malignant tumors (1). Lung cancer is classified by non-small cell lung cancer and small cell lung cancer, and lung adenocarcinoma is the most common type of non-small cell lung cancer (2). Lung adenocarcinoma originates from bronchial mucosa epithelium and bronchial mucosa. Due to the difficulty of early diagnosis and rapid spread of the cancer cells, lung adenocarcinoma has a high mortality (2). Therefore, it is of significance to study the potential markers of lung adenocarcinoma and to inhibit the proliferation and migration of cancer cells.
Seven subunits, including Coatomer protein complex subunit β (COPB2), are contained in coatomer protein complex I (COPI). The molecular weight of COPB2 is 102 kD, and encoded β'-COP contains 906 amino acid residues of nuclear protein (3). Research (4-6) proved that COPB2 is overexpressed in various cancer cells such as colon cancer, prostate cancer, cervical cancer, gastric cancer, and is closely related to the occurrence and development of tumors.
MicroRNAs (miRNAs) are a type of non-coding small RNAs with approximately 18-25 nucleotides in length and are encoded by endogenous genes that regulate mRNA degradation or block mRNA translation through binding to the 3' untranslated region (UTR) of the target mRNA (7,8). miRNAs are involved in many biological processes such as cell differentiation, reproduction, metabolism, and apoptosis, and they are potential targets for tumor intervention treatment. Xu et al. (9) demonstrated that miR-502 promotes tumor overgrowth in esophageal cancer by promoting phosphorylation of serine/threonine kinase (AKT) pathway; Cagle et al. (10) showed that miR-214 can down-regulate protein tyrosine kinase 6 (PTK6) and inhibit the development of prostate cancer cells. Researchers also found that miR-203a-3p targeting ATM gene inhibits the proliferation of ovarian cancer cells and promotes apoptosis of the cancer cells (11).
There is little published data on COPB2 in lung cancer and lung adenocarcinoma. Our previous experiments found that COPB2 was overexpressed in lung adenocarcinoma cells. The current study investigated the mechanism of COPB2 in lung adenocarcinoma cells, aiming to provide a new basis for the treatment of lung adenocarcinoma.
Methods
Cell culture
Normal lung epithelial cells BEAS-2B (95102433-RNA-5UG, SIGMA,USA) and lung adenocarcinoma cell lines H1299 (CRL-5803, ATCC, USA), A549 (KG007, KeyGEN BioTECH, China), SK-MES-1 (KG153, KeyGEN BioTECH, China), NCI-H23 (CM-0397, Procell, China), H1975 (CX0373, Boster, China) were purchased from the Chinese Academy of Sciences Cell Bank Type Library (Shanghai, China). The cells were separately added into Roswell Park Memorial Institute-1640 (RPMI-1640, PM150110, Procell, China) medium containing 10% fetal calf serum (FBS), and then cultured in a cell incubator containing 5% CO2 at 37 °C. The medium were pre-added with 100 U/mL penicillin and 100 µg/mL streptomycin.
Small interfering RNA and cell transfection
Plasmid for COPB2 gene (CAT#: SC117165), a small interfering RNA (siRNA) to COPB2, and a negative control siRNA (CAT#: SR30004) were purchased from OriGene (USA). SiRNA sequences targeting COBP2 were designed according to one previous research (12). One day before transfection, the cells were trypsinized and seeded in antibiotic-free RPMI-1640 medium containing 10% FBS. When 50% confluence was reached, the cells were washed by PBS and placed in OPTI-MEM medium (31985062, Invitrogen, USA). Thirty pmol miR-216a-3p mimic or inhibitor and 1 µg DNA were separately diluted by 50 µL OPTI-MEM medium. According to the instructions of Lipofectamine 2000 Transfection Reagent (Cat. No. 11668-027, Invitrogen, USA), 1 µL Lipofectamine 2000 solution was diluted by 50 µL OPTI-MEM medium, and mixed together. The medium was changed after incubation at 37 °C for 5 hours (h). After 24 h, the transfection efficiency was measured by qRT-PCR.
Total RNA extraction and quantitative real time-polymerase chain reaction (qRT-PCR)
Total RNAs in each cell line were extracted by Trizol reagent. After detecting the concentration of RNA by a UV Spectrophotometer (NanoDrop2000, Thermo Scientific, USA), total RNAs were then synthesized using Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622, USA). Glyceraldehyde-3-phosphatede hydrogenase (GAPDH) served an internal reference. The primer sequences used in qRT-PCR were as follows: COPB2-F, 5'-CTTCCTGTTCGAGCTGCAAAG-3', and COPB2-R, 5'-CACTCTAATCTGCATGTCATCCG-3'; GAPDH-R, 5'-GGCTGTTGTCATACTTCTCATGG-3', and GAPDH-F, 5'-GGAGCGAGATCCCTCCAAAAT-3'. According to the manufacturer's instructions, miR-216a-3p was detected using miRNA reverse transcription kit (QP013, GeneCopoeia, USA) and TaqMan® Universal PCR Master Mix (4304437, ABI, USA). U6 served as an internal control. The primer sequences were as follows: miR-216a-3p-F, 5'-GCTGGCAACTGTGAGTCGTA-3', and miR-216a-3p-R, 5'-CTCAGCCGTTAACGTGACCT-3'; U6-F, 5'-AAAGCAAATCATCGGACGACC-3', and U6-R, 5'-GTACAACACATTGTTTCCTCGGA-3'. Ten µL FastStart Universal SYBR Green Master (04913914001, Roche, Switzerland), 6 µL diethyl pyrocarbonate water (DEPC water, R1600, Solarbio, China), 2 µL diluted cDNA, and 1 µL of each diluted upstream and downstream primers were added to a 96-well plate and mixed together. The reaction system was conducted on a PCR machine (C1000 Thermal Cycler, BIO RAD, USA) under the following amplification conditions: pre-denaturation at 95 °C for 10 minutes (min), denaturation at 95 °C for 15 seconds (sec), annealing at 60 °C for 1 min, for a total of 40 cycles. The gene expression level was calculated by2−ΔΔCT.
Western blotting
The cells were washed by PBS once, digested by 0.025% trypsin (P822466-100g, Macklin, China), and then rinsed in PBS and collected into a centrifugal tube. One hundred µL RIPA lysatebuffer (P0013C, Beyotime, China) was added to the cells and blown evenly. The cells were centrifuged (1-15PK, Sigma, USA) at 4 °C, 16,000 ×g, and placed in a centrifugal tube. The protein concentration was determined by bicinchoninic acid (BCA) method, 100 µg protein was separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (MILLIPORE, IPVH00010, USA). The membrane was washed by the Tris Buffered Saline and Tween 20 (TBST) 3 times (50 mM Tris, 150 mM NaCl and 2% Tween-20, pH 7.5) at room temperature for 10 min. Cleaved Caspase-3 (ab2302, 1:1,000, abcam, UK), Caspase-3 (ab32351, 1:5,000, abcam, UK), BCL2 apoptosis regulator (Bcl-2, ab59348, 1:1,000, abcam, UK), BCL2 associated X (Bax, ab32503, 1:5,000 abcam, UK), E-Cadherin (ab40772, 1:10,000, abcam, UK), N-Cadherin (ab18203, 1:1,000, abcam, UK), Vimentin (ab92547, 1:2,000, abcam, UK), GAPDH (ab8245, 1:20,000, abcam, UK) antibodies and corresponding bands were incubated overnight at 4 °C. On the next day, the PVDF membrane was washed 3 times by TBST at room temperature for 10 min and then incubated with corresponding secondary antibodies (Anti-mouse IgG, #7076, 1:2,000, Cell Signaling Technology; Anti-rabbit IgG, #7074, 1:2,000, Cell Signaling Technology) for 1 h at room temperature. The PVDF membrane was washed by TBST 3 times. The protein bands were detected by ECL (RPN2232, GE Amersham, USA) in Gel imager and imaging system (Universal Hood II, BIO RAD, USA). Finally, the images were analyzed using Image J software (1.48, National Institutes of Health, USA). GAPDH served as an endogenous control.
Cell viability
Cell suspension (100 µL) was placed in a 96-well plate and cultured in an incubator for 24 h at 37 °C in 5% CO2. Each group of cells was then seeded into a culture plate at 5×103 per well. After incubating for 0, 24, 48, and 72 h, 10 µL Cell Couting Kit-8 (CCK-8, YZ-CK04-500T, Solarbio, China) solution was added. After incubating for 1 h in the incubator, cell absorbance was measured at 450 nm by a microplate reader (SpectraMax i3x, Molecular Devices, USA). Finally, cell viability was determined using GraphPad Prism software version 6.0.
Flow cytometry
The effects of changes of COPB2 expression on apoptosis were examined by flow cytometry. Cells at the log phase were seeded in a medium at a density of 5×105 cells per well, and incubated in a 6-well plate for 24 h. The cells were then washed in phosphate buffered saline (PBS), centrifuged at 1,000 r/min for 5 min. Five µL Annexin V-FITC (130-093-060, Miltenyi, Germany) was added to the cells, gently mixed and incubated for 15 min, the supernatant was discarded, and the cells were added with 10 µL propidium iodide (PI, JBS-LI-001, ENZO, USA). Finally, the apoptosis of cells were analyzed by flow cytometry (FCM, 322457, Bio-Rad, USA).
Cell migration and invasion
Cell migration and invasion were detected by Transwell assay. The cells were fasted for 24 h and then trypsinized in 0.1% FBS supplementation medium. Transwell chambers (8.0 µm pore size, 353097-1, FALCON, USA) were placed in the plates, and 10% FBS medium was added to the plates to provide chemoattractants. The upper and lower chambers were separated by a polycarbonate membrane (8.0 µm pore size, 353097-1, FALCON, USA). For detection of cell invasion, cells (1×104 cells per well) in serum-free medium were seeded onto the upper chamber which was pre-coated with Matrigel on the bottom. After 48 h, matrigel in the Transwell chamber was removed using cotton swab. The cells on the basal side of the membrane were first fixed by 4% paraformaldehyde for 10 min and then stained by 0.1% crystal violet stain (G1063-10, Solarbio, China) for 20 min at room temperature. Finally, the cells were observed under a fluorescent inverted microscope (IX71, OLYMPUS, Japan) and counted.
TargetScan forecast
TargetScan database (http://www.targetscan.org) was used to predict target genes. To explore the relationship between miR-216a-3p and COBP2 in lung adenocarcinoma cells was predicted by TargetScan database.
Luciferase assay
A luciferase reporter plasmid (AP-MN-P-50, Axygen, USA) containing the COPB2-3'-UTR sequence and the mutant COBB2-3'-UTR (COPB2-3'-UTR-mut) sequence were respectively constructed. MiR-216a-3p mimics (CMH1914, Cohesion Bio, UK) could enhance endogenous miR-216a-3p and was used in the following experiments. The cells were seeded at a density of 50% in a 12-well plate containing DMEM medium (C11995500BT, Invitrogen, USA). After incubation for 16 h, the COPB2-3'-UTR or COPB2-3'-UTR-mut plasmid was co-transfected with miR-216a-3p mimic or control mimic, respectively. The medium was changed 6 h after the transfection. After cell transfection for 24 h at 37 °C, the luciferase assay was detected by dual luciferase kit (Invitrogen, 1168-019, USA). 5× lysate (RIPA) was mixed with sterile water at a ratio of 1:4. After washing the cells by PBS, the cells were lysed using the prepared lysate and then incubated for 15 min at room temperature. Twenty µL lysed cells were added with 100 µL LAR II reagent to detect the firefly fluorescence value by GloMax 2020 (YQ1633114524, Promega, USA). Then, 100 µL Stop &Glo reagent was then added to the cells to detect Renilla fluorescence (Renilla was an internal reference). Finally, the ratio of firefly/Renilla was calculated.
Statistical analysis
Statistical analysis was performed by SPSS 20.0 (SPSS Inc., Chicago, IL, USA) and the data were expressed as the mean ± standard deviation (SD). Comparisons among groups were performed using ANOVA. Differences were determined by the Student two-tailed t-test between two groups. P<0.05 was considered to be statistically significant.
Results
COPB2 was high-expressed in lung adenocarcinoma cells
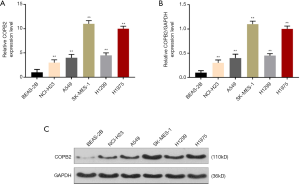
COPB2 expression was analyzed by qRT-PCR and Western blotting. The results showed that COPB2 was high-expressed in various lung adenocarcinoma cells (P<0.001, Figure 1A,B,C), and its expression was the highest in SK-MES-1 cells. Therefore, SK-MES-1 cells were used in the following experiments.
SiCOPB2 promoted tumor cell apoptosis and inhibited cell viability, migration and invasion
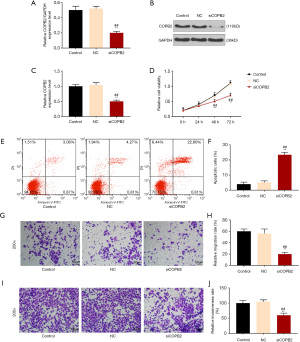
SiCOPB2 cells were obtained by transfection technique, and then the transfection was examined by Western blotting and qRT-PCR. The results showed that the expression of COPB2 was significantly reduced in the siCOPB2 cells (P<0.001, Figure 2A,B,C). The results of CCK-8 showed that the cell viability of the siCOPB2 group was lower than that of the control group (P<0.001, Figure 2D). Flow cytometry results showed that apoptosis of the cells in siCOBP2 group was greatly increased, which was statistically different from the control group (P<0.001, Figure 2E,F). By comparing the cell migration and invasion ability, it was found that cell number in the siCOBP2 group in the visual field was significantly reduced, but lower than that of the control group (P<0.001, Figure 2G,H,I,J). The transfected siCOPB2 cells inhibited the expression of COPB2 gene in tumor cells. By detecting cell viability, migration, invasion and apoptosis, we found that siCOPB2 promoted apoptosis and inhibited cell migration and invasion. The above results indicate that COPB2 can promote the proliferation and growth of tumor cells and inhibit apoptosis.
MiR-216a-3p could target COBP2
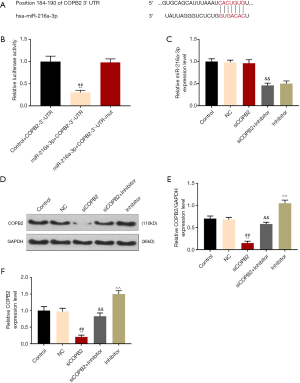
TargetScan database predicted that the COPB2 gene has a miR-216a-3p binding site at its 3'UTR (Figure 3A), suggesting that COPB2 expression could be regulated by miR-216a-3p. The luciferase assay showed that the expression of luciferase in the experimental group was lower than that in the control group (P<0.001, Figure 3B). QRT-PCR was performed to detect the mRNA expression level of miR-216a-3p (Figure 3C). No significant difference was observed between siCOPB2 group and negative control group, indicating that COPB2 has no regulatory effect on miR-216a-3p. Furthermore, qRT-PCR and Western blotting were performed to verify whether miR-216a-3p regulated the expression of mRNA and protein levels of COPB2 genes. The results showed that the expression of COPB2 was up-regulated in miR-216a-3p inhibitor group (compared with the siCOPB2 group, P<0.001, Figure 3D,E,F), suggesting that miR-216a-3p could down-regulated the expression of COPB2. The above experimental results indicated that miR-216a-3p could down-regulating the expression of COPB2 gene in tumor cells.
The effect of SiCOPB2 on promoting apoptosis of lung adenocarcinoma cells and inhibiting cell metastasis could be partially reversed by miR-216a-3p inhibitor
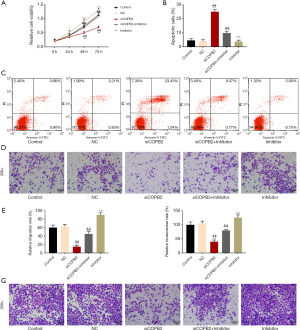
The results of CCK8 assay showed that the cell viability of miR-216a-3p inhibitor was significantly enhanced compared with the siCOPB2 group (Figure 4A), and flow cytometry results showed that miR-216a-3p inhibitor reduced apoptosis of cancer cells induced by siCOPB2 (P<0.001, Figure 4B,C). The results of cell migration and invasion experiments revealed that compared with the siCOPB2 group, the migration and invasion abilities of miR-216a-3p inhibitor group were improved, and the number of cells in the experiment was significantly increased (P<0.001, Figure 4D,E,F,G). The above results suggest that miR-216a-3p inhibitor can promote cell migration and invasion, inhibit the apoptosis induced by siCOPB2. The results further confirmed that miR-216a-3p can promote the apoptosis of lung adenocarcinoma cells and inhibit the proliferation and migration of cancer cells through down-regulating the expression of COPB2 gene in tumor cells.
Effects of COPB2 on apoptosis and migration of the cancer cells
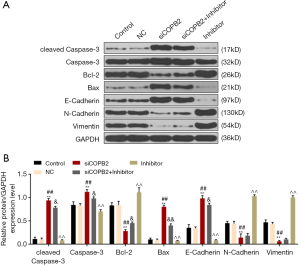
As shown in Figure 5, silencing COBB2 significantly up-regulated the expressions of cleaved Caspase-3, Caspase-3, Bax and E-Cadherin protein levels (P<0.001), while the expressions of Bcl-2, N-Cadherin and Vimentin was inhibited (P<0.001, Figure 5A,B), indicating that silencing COPB2 promoted apoptosis and inhibited metastasis of cancer cells, however, the miR-216a-3p inhibitor reversed such effects. In cells transfected with siCOPB2 and miR-216a-3p inhibitor, cleaved Caspase-3, Caspase-3, Bax and E-Cadherin were down-regulated, and the expression of Bcl-2 was increased, while N-Cadherin and Vimentin did not change significantly. The results showed that miR-216a-3p inhibitor can partially reverse siCOPB2 to promote tumor cell apoptosis and inhibit migration.
Discussion
The current study revealed the targeted relationship between COPB2 and miR-216a-3p, and their important roles in the development of lung adenocarcinoma. COPB2 was high-expressed in lung adenocarcinoma cells, and after silencing COBB2, proliferation and invasion of lung adenocarcinoma cells were significantly inhibited, by contrast, the apoptosis of cancer cells was greatly increased. COPB2 (3,13) plays an important role in the process of transporting intracellular protein and endoplasmic reticulum Golgi bubble-mediated. Scientists found that down-regulating COPB2 expression can promote cancer cell apoptosis (14-16), which confirmed our experimental results. In addition, miR-216a-3p was predicted as target gene of COPB2, and its inhibitor partially reversed the intervention of siCOPB2 on lung adenocarcinoma cells. To the best of our knowledge, we were the first to prove that miR-216a-3p can down-regulate COPB2, thereby inhibiting the migration and promoting apoptosis of cancer cells.
More than 20,000 miRNAs have been discovered since its discovery in 2000 (17). MiRNAs account for about 1–3% of human genes and regulate transcription of at least half of human genes, moreover, they can not only regulate target genes at the transcriptional level, but also at translation levels. As endogenous coding RNAs, miRNAs can also regulate genes involved in development of tumor, and they enter tumor cells more easily than siRNAs (18-20). The synthesis of miRNAs can regulate cell homeostasis and control the expressions of cancer-related enzymes. MiRNAs in body fluids can promote cell-to-cell connections. Some miRNAs with distinctive characteristics can be used for the diagnosis of different diseases. In addition, single-stranded miRNAs can interfere with multiple targets in a pathway. Studies confirmed that miRNAs and cancer cells are related to cell cycle, apoptosis and metastasis, suggesting a regulatory role in the development of cancer (21-23). The current study was the first to demonstrate that silencing COBB2 could inhibit metastasis and invasion of lung adenocarcinoma cells and promote apoptosis. Then, mir-216a-3p targeting COPB2 was verified, and found to have similar effects to siCOPB2, indicating that mir-216a-3p can replace siCOPB2 and directly down-regulate COPB2 expression in vivo. Moreover, mir-216a-3p avoids the physiological barrier that siCOPB2 encounters during its propagation in the body, preventing itself from entering the tumor cells (24,25).
Lung adenocarcinoma occurs in bronchioles and alveolar epithelial cells (26). Finding regulatory genes for lung adenocarcinoma, inhibiting cancer cell metastasis, and accelerating cancer cell apoptosis are the key to the treatment of the disease (27). E-Cadherin and N-Cadherin are important components of cadherin and an important structure for adhesion between cells (28,29). Studies showed that (30), E-Cadherin and N-Cadherin play important roles in tumor metastasis and invasion. The conversion of E-Cadherin to N-Cadherin is one of the important mechanisms of epithelial-mesenchymal transition (EMT) and a potential mechanism for tumor development and metastasis. Vimentin is an intermediate fibrin in mesenchymal cells and skeleton protein that maintains cell integrity (31), more importantly, it is involved in cell adhesion, migration and apoptosis, and is high-expressed in a variety of tumors (32). We confirmed by Western blotting that silencing COPB2 can significantly up-regulate E-Cadherin and inhibit the expressions of N-Cadherin and Vimentin, thereby inhibiting the migration and invasion of lung adenocarcinoma cells. The mir-216a-3p inhibitor can only inhibit the anti-metastatic effect of siCOPB2 by down-regulating E-Cadherin.
Through apoptosis, cells actively maintain cell life in order to maintain homeostasis, eliminate useless or cancerous cells (33), therefore, apoptosis of cancer cells is seen to be able to protect normal tissues and treat cancers through inhibiting cancer cell proliferation (34,35). Resveratrol (36), bleomycin (37), dexamethasone (38) and other chemotherapeutic drugs, which could induce apoptosis of cancer cells, are widely used to treat cancers, and Caspase-3, Bax and Bcl-2 are key proteins regulating apoptosis. Bax and Bcl-2 belong to the Bcl-2 family, but the difference is that Bcl-2 is an anti-apoptotic protein, while Bax is a pro-apoptotic protein (39). In addition to the Bcl-2 family, the Caspase family also plays a key role in the process of apoptosis. Caspase-3 is one of the important apoptosis-executing proteins (39). Caspase-3 is a downstream regulatory protein of Bax and Bcl-2 (39,40). Bcl-2 can prevent the release of cytochrome C, thereby blocking the activation of Caspase protease and ultimately inhibiting apoptosis. Bax help cytochrome C enter the mitochondria and further activate Caspase-3 to promote apoptosis. Our study showed that cleaved Caspase-3, Caspase-3 and Bax proteins were up-regulated and Bcl-2 expression was down-regulated in lung adenocarcinoma cells through silencing COBP2. However, the effects of mir-216a-3p inhibitor on siCOPB2 were reversed, pro-apoptotic protein expression was inhibited, and anti-apoptotic protein expression was up-regulated, moreover, these results were consistent with our previous observations on cell apoptosis of siCOPB2 cells.
Conclusions
In conclusion, the present study confirms that silencing COPB2 promotes apoptosis and inhibits cell metastasis of lung adenocarcinoma cells, and such effects can be enhanced by miR-216a-3p inhibitor. Moreover, the result demonstrates for the first time that the intervention mechanism of miR-216a-3p underlying lung adenocarcinoma is related to the up-regulation of cleaved Caspase-3, Caspase-3, Bax, E-Cadherin and inhibition of Bcl-2 expression. This study provides new intervention targets for miRNA treatment of lung adenocarcinoma.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.65). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- De Baere E, Speleman F, Van Roy N, et al. Assignment of the cellular retinol-binding protein 1 gene (RBP1) and of the coatomer beta subunit gene (COPB2) to human chromosome band 3q23 by in situ hybridization. Cytogenet Cell Genet 1998;82:226-7. [Crossref] [PubMed]
- Mi Y, Sun C, Wei B, et al. Coatomer subunit beta 2 (COPB2), identified by label-free quantitative proteomics, regulates cell proliferation and apoptosis in human prostate carcinoma cells. Biochem Biophys Res Commun 2018;495:473-80. [Crossref] [PubMed]
- Tan MS, Chang SW, Cheah PL, et al. Integrative machine learning analysis of multiple gene expression profiles in cervical cancer. Peer J 2018;6:e5285. [Crossref] [PubMed]
- Li ZS, Liu CH, Liu Z, et al. Downregulation of COPB2 by RNAi inhibits growth of human cholangiocellular carcinoma cells. Eur Rev Med Pharmacol Sci 2018;22:985-92. [PubMed]
- Voglova K, Bezakova J, Herichova I. Progress in micro RNA focused research in endocrinology. Endocr Regul 2016;50:83-105. [Crossref] [PubMed]
- Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov 2016;6:235-46. [Crossref] [PubMed]
- Xu J, Pan X, Hu Z. MiR-502 mediates esophageal cancer cell TE1 proliferation by promoting AKT phosphorylation. Biochem Biophys Res Commun 2018;501:119-23. [Crossref] [PubMed]
- Cagle P, Niture S, Srivastava A, et al. MicroRNA-214 targets PTK6 to inhibit tumorigenic potential and increase drug sensitivity of prostate cancer cells. Sci Rep 2019;9:9776. [Crossref] [PubMed]
- Liu HY, Zhang YY, Zhu BL, et al. MiR-203a-3p regulates the biological behaviors of ovarian cancer cells through mediating the Akt/GSK-3beta/Snail signaling pathway by targeting ATM. J Ovarian Res 2019;12:60. [Crossref] [PubMed]
- Pu X, Wang J, Li W, et al. COPB2 promotes cell proliferation and tumorigenesis through up-regulating YAP1 expression in lung adenocarcinoma cells. Biomed Pharmacother 2018;103:373-80. [Crossref] [PubMed]
- Presley JF, Ward TH, Pfeifer AC, et al. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature 2002;417:187-93. [Crossref] [PubMed]
- Wang Y, Chai Z, Wang M, et al. COPB2 suppresses cell proliferation and induces cell cycle arrest in human colon cancer by regulating cell cycle-related proteins. Exp Ther Med 2018;15:777-84. [PubMed]
- An C, Li H. Zhang Xet al. Silencing of COPB2 inhibits the proliferation of gastric cancer cells and induces apoptosis via suppression of the RTK signaling pathway. Int J Oncol 2019;54:1195-208. [PubMed]
- Bhandari A, Zheng C, Sindan N, et al. COPB2 is up-regulated in breast cancer and plays a vital role in the metastasis via N-cadherin and Vimentin. J Cell Mol Med 2019;23:5235-45. [Crossref] [PubMed]
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. [Crossref] [PubMed]
- Backes C, Meese E, Keller A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol Diagn Ther 2016;20:509-18. [Crossref] [PubMed]
- Del Vescovo V, Denti MA. microRNA and Lung Cancer. Adv Exp Med Biol 2015;889:153-77. [Crossref] [PubMed]
- Grisard E, Nicoloso MS. Following MicroRNAs Through the Cancer Metastatic Cascade. Int Rev Cell Mol Biol 2017;333:173-228. [Crossref] [PubMed]
- Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts 2017;8:61-81. [Crossref] [PubMed]
- Sartorius K, Sartorius B, Winkler C, et al. The biological and diagnostic role of miRNA's in hepatocellular carcinoma. Frontiers in bioscience (Landmark edition) 2018;23:1701-20.
- Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol 2016;98:12-23. [Crossref] [PubMed]
- Lee SJ, Son S, Yhee JY, et al. Structural modification of siRNA for efficient gene silencing. Biotechnol Adv 2013;31:491-503. [Crossref] [PubMed]
- Suter SR, Sheu-Gruttadauria J, Schirle NT, et al. Structure-Guided Control of siRNA Off-Target Effects. J Am Chem Soc 2016;138:8667-9. [Crossref] [PubMed]
- Ding F, Wang D, Li XK, et al. Overexpression of S100A14 contributes to malignant progression and predicts poor prognosis of lung adenocarcinoma. Thorac Cancer 2018;9:827-35. [Crossref] [PubMed]
- Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e1S-e29S.
- Petrova YI, Schecterson L, Gumbiner BM. Roles for E-cadherin cell surface regulation in cancer. Mol Biol Cell 2016;27:3233-44. [Crossref] [PubMed]
- Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. The International journal of developmental biology 2004;48:463-76. [Crossref] [PubMed]
- van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nature reviews Cancer 2014;14:121-34. [Crossref] [PubMed]
- Costigliola N, Ding L, Burckhardt CJ, et al. Vimentin fibers orient traction stress. Proc Natl Acad Sci U S A 2017;114:5195-200. [Crossref] [PubMed]
- Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011;68:3033-46. [Crossref] [PubMed]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007;35:495-516. [Crossref] [PubMed]
- Shivapurkar N, Reddy J, Chaudhary PM, et al. Apoptosis and lung cancer: a review. J Cell Biochem 2003;88:885-98. [Crossref] [PubMed]
- Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin 2005;55:178-94. [Crossref] [PubMed]
- Rauf A, Imran M, Butt MS, et al. Resveratrol as an anti-cancer agent: A review. Crit Rev Food Sci Nutr 2018;58:1428-47. [Crossref] [PubMed]
- Chiani M, Azadmanesh K, Shokrgozar MA, et al. Enhanced antitumor effect of targeted nanoliposomal bleomycin. Chem Biol Drug Des 2017;90:953-61. [Crossref] [PubMed]
- Wang LJ, Lu W, Zhou TY. Current applications of dexamethasone for cancer treatment. Yao Xue Xue Bao 2015;50:1217-24. [PubMed]
- Hassan M, Watari H, AbuAlmaaty A, et al. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int 2014;2014:150845.
- Babu PP, Suzuki G, Ono Y, et al. Attenuation of ischemia and/or reperfusion injury during myocardial infarction using mild hypothermia in rats: an immunohistochemical study of Bcl-2, Bax, Bak and TUNEL. Pathol Int 2004;54:896-903. [Crossref] [PubMed]


