
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-2020-006
Correlation among VEGFR3 gene promoter methylation, protein overexpression, and clinical pathology in early gastric cancer
Introduction
Gastric cancer, one of the most prevalent malignant tumors and the third leading cause of cancer mortality worldwide (1), is divided into early gastric cancer (EGC) and advanced gastric cancer. EGC is characterized as gastric cancer with limited aggressiveness within the submucosa, and accounts for major incident gastric cancer in Eastern Asia. Although the 5-year postoperative survival rate of EGC exceeds 90%, a poor prognosis is correlated with lymph node metastasis. Therefore, a better understanding of the role of molecular biomarkers in EGC metastasis is important. Vascular endothelial growth factor receptor 3 (VEGFR3) is a lymphatic endothelial biomarker also found in blood vessels and malignant tumor cells, indicating its involvement in cancer lymphangiogenesis and metastasis (2,3). The occurrence and development of EGC are cumulative processes of multiple factors, stages, and genes. Both genetic and epigenetic mechanisms play important roles in the molecular mechanism of gastric cancer. DNA methylation is one of the most extensively studied epigenetic processes and the methylation degree of certain genes can be an efficient biomarker for early detection, evaluating prognosis and disease recurrence, and even targeted therapy (4-6). Digestive tract cells have a high degree of abnormal methylation and tumor susceptibility (7).
In this study, methylation status of the VEGFR3 gene promoter and protein overexpression in the EGC tissue of 50 patients were examined by methylation-specific PCR (MSP) and immunohistochemistry. We also investigated the correlation between VEGFR3 gene methylation and its protein expression, as well as the correlation between abnormal methylation of the VEGFR3 gene promoter and clinicopathological factors of EGC. The results of this study provide insights into whether the VEGFR3 epigenetic biomarker can be used for early diagnosis and prognosis in EGC patients.
Methods
Sample collection
This study included specimens of 50 EGC cases from patients admitted to Qilu Hospital of Shandong University (Shandong, China) between 2013 and 2014. The study population included 27 males and 23 females, with a median age of 54 years (14 patients ≥60 years of age, 36 patients <60 years of age). Two senior pathologists reviewed and interpreted the routine pathological sections according to the 2006 World Health Organization tumor classification and diagnostic criteria (8). Among these 50 cases, there were 20 cases of highly and moderately differentiated adenocarcinoma and 30 cases of poorly differentiated adenocarcinoma including signet ring cell carcinoma and mucinous adenocarcinoma; 30 cases of intramucosal carcinoma and 20 cases of submucosal carcinoma; 12 cases with lymph node metastasis and 38 cases without lymph node metastasis. At the same time, 20 cases of normal gastric mucosal tissues at the incised margin of EGC were selected as the control group. None of the patients received anti-tumor treatment such as radiotherapy and/or chemotherapy before surgery, and the clinical data were complete. This study was approved by the Regional Ethics Committee of Qilu Hospital of Shandong University. Because this was a retrospective study based on the medical records and tumor slides assessment, the written informed consent requirement was waived.
Immunohistochemistry staining
In each case, the primary and gastric mucosal tissue of the incised margin were excised, embedded into wax blocks, and sectioned into 3 µm thick slides. After being pretreated with 3% hydrogen peroxide at room temperature for 5–10 min, the slides were placed in antigen repair solution (ZLI-9064; Zsbio Commerce Store, Beijing, China), followed by microwave incubation. Then the sections were incubated with mouse anti-human VEGFR3 monoclonal antibody (1:100, ZM-0277; Zsbio Commerce Store, Beijing, China) overnight at 4 °C after being blocked in goat serum. The slides were further washed and incubated with biotinylated secondary antibody and horseradish-labeled streptavidin working solution (SP-9001, SP-9002; Zsbio Commerce Store, Beijing, China) at room temperature for 2 h. The stained tissues were finally developed in DAB colorant (ZLI-9018; Zsbio Commerce Store, Beijing, China) and the slides were counterstained with hematoxylin.
Methylation-specific polymerase chain reaction
For DNA extraction, about 10 pieces of each tissue wax blocks of 3 µm were sliced and placed in a 1.5 mL Eppendorf tube. The DNA of paraffin-embedded tissue samples was extracted with the paraffin genomic DNA extraction kit (QIAGEN Hilden, Germany). Sulfite modification of quantified DNA was conducted with a methylation modification kit (Merck Millipore, Hayward, CA, USA). VEGFR3 methylation-specific primer pairs (5'-GTC GGT TAT TTC GGG TGT TTC-3' and 5'-AAT ATC GAC GAA CAA TAT CGA CG-3') and non-methylation primer pairs (5'-GGG TTG GTT ATT TTG GGT TT-3' and 5'-ACA CAA TAT CAA CAA ATA TCA ACA-3') were synthesized by Shanghai Sangon Biological Company (Shanghai, China). PCR reactions were performed under the following conditions: 95 °C pre-denaturation for 10 min, 94 °C denaturation for 30 s, 45 °C annealing for 30 s, 72 °C extension for 45 min, and 72 °C extension for 10 min. Electrophoresis was performed and a photograph was taken using a gel imaging system (JY04S-3C; Beijing Junyi, Beijing, China).
Data analysis
For immunohistochemistry analysis, positive cells with brown-yellow substances in the cytoplasm were observed and counted under a light microscope. The sections with <10% positive cells were defined as negative (−), 10–25% positive cells were defined as weakly positive (+), 26–50% positive cells were defined as medium positive (++), and >50% positive cells were defined as strongly positive (+++). SPSS 11.0 statistical software was used to perform the chi-square test and Fisher’s exact probability test. P<0.05 was considered statistically significant.
Results
Expression of VEGFR3 in EGC
Positive VEGFR3 expression in granules in EGC was stained brown-yellow and predominantly found in the tumor cell cytoplasm (Figure 1A,B,C,D). The positive expression of VEGFR3 in 50 EGC tissues was 60%, which was significantly higher than that in normal gastric mucosal tissues (10%) (P<0.05; Table 1). In addition, VEGFR3 was overexpressed in 11 of 12 tumors with lymph node metastasis, which was statistically different from the 19 of 38 tumors without lymph node metastasis (P<0.05). In contrast, no statistical difference in VEGFR3 expression was found among cases with different gender, age, tumor size, differentiation degree, or depth of tumor infiltration (P>0.05). Taken together, these results suggest that VEGFR3 expression is significantly correlated with the presence of EGC malignant tissue with lymph node metastasis.
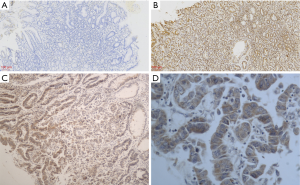
Table 1
| Clinical parameters | Case | Present | Absent | P |
|---|---|---|---|---|
| Sex | ||||
| Male | 27 | 16 | 11 | |
| Female | 23 | 14 | 9 | >0.05 |
| Patient age | ||||
| <60 | 36 | 20 | 16 | |
| ≥60 | 14 | 10 | 4 | >0.05 |
| Tumor size | ||||
| ≤2 cm | 21 | 10 | 11 | |
| >2 cm | 29 | 20 | 9 | >0.05 |
| Tumor | ||||
| High-middle grade | 20 | 11 | 9 | |
| Low grade | 30 | 19 | 11 | >0.05 |
| Depth of invasion | ||||
| Mucosal | 30 | 14 | 16 | |
| Submucosal | 20 | 16 | 4 | <0.05 |
| Lymph node metastasis | ||||
| Without | 38 | 19 | 19 | |
| With | 12 | 11 | 1 | <0.05 |
Methylation status of the VEGFR3 promoter in EGC
The MSP method was used to detect DNA methylation in the promoter of the VEGFR3 gene using genomic DNA template modified with sodium bisulfite. The electrophoresis results are shown in Figure 2. To analyze the MSP results, the electrophoretic bands of the samples were compared with the DNA ladder bands and the obviously weaker bands were described as negative. Methylation was detected in 24 of the 50 EGC patients (48.0%) and 17 of the 20 normal gastric mucosa (85%). The methylation frequency of VEGFR3 gene in gastric cancer was significantly lower than that in the normal gastric mucosa (P<0.05; Table 2).
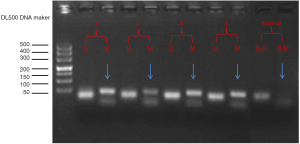
Table 2
| Promoter methylation | Cases | VEGFR3 | P | |
|---|---|---|---|---|
| + | − | |||
| Unmethylation | 26 | 20 | 6 | |
| Methylation | 24 | 10 | 14 | <0.05 |
Correlation between methylation of VEGFR3 gene promoter and protein expression in EGC
As shown in Table 2, among the 30 cases of gastric cancer with positive VEGFR3 expression, 20 had no methylation detected by MSP (66.67%). In addition, among the 20 cases of gastric cancer with negative VEGFR3 expression, 14 had MSP detectable methylation (70%). These results indicate that overexpression of VEGFR3 protein is closely related to hypomethylation of the VEGFR3 gene promoter in EGC tumor cells.
Correlation between methylation status of VEGFR3 gene promoter and clinicopathological characteristics of EGC patients
As shown in Table 3, 22 of the 38 EGC tumor samples without lymph node metastasis showed detectable methylation, whereas 2 of the 12 samples with lymph node metastasis showed hypomethylation. There was a statistical difference between the methylation detectable rate of VEGFR3 gene and lymph node metastasis of EGC cases (P<0.05). By contrast, no statistical difference in VEGFR3 promoter methylation status was found among cases with different gender, age, differentiation degree, or depth of tumor infiltration (P>0.05). Taken together, these results suggest that hypomethylation of VEGFR3 gene promoter is significantly correlated with the presence of EGC malignant tissue with lymph node metastasis.
Table 3
| Clinical parameters | Case | VEGFR3 methylation | P | |
|---|---|---|---|---|
| Present | Absent | |||
| Sex | ||||
| Male | 27 | 16 | 11 | |
| Female | 23 | 8 | 15 | >0.05 |
| Patient age | ||||
| <60 | 36 | 18 | 18 | |
| ≥60 | 14 | 6 | 8 | >0.05 |
| Tumor size | ||||
| ≤2 cm | 21 | 11 | 10 | |
| >2 cm | 29 | 13 | 16 | >0.05 |
| Tumor | ||||
| High-middle grade | 20 | 9 | 11 | |
| Low grade | 30 | 15 | 15 | >0.05 |
| Depth of invasion | ||||
| Mucosal | 30 | 14 | 16 | |
| Submucosal | 20 | 10 | 10 | >0.05 |
| Lymph node metastasis | ||||
| Without | 38 | 22 | 16 | |
| With | 12 | 2 | 10 | <0.05 |
Discussion
VEGFRs are members of the receptor tyrosine kinase superfamily. They contain an extracellular ligand-binding domain consisting of seven immunoglobulin-like folds, a single transmembrane region, and a split tyrosine kinase domain. VEGFRs activate the mitogen-activated protein kinase and AKT signaling pathways in the cytoplasm to promote cell proliferation and migration (9-12). As a lymphangia-specific growth factor receptor, in normal adult tissues, VEGFR3 is mainly expressed in lymphoid endothelial cells, but not in vascular endothelial cells. Previous studies have revealed that the growth of new lymphatic vessels in tumors is mainly achieved through the regulation of VEGFR3 located on lymphatic endothelial cells. Vascular endothelial growth factor C (VEGFC) secreted by tumor cells specifically binds to its receptor VEGFR3, which causes the lymphatic vessel germ around the tumor to grow into tumor tissue. Studies have shown that the VEGF-C-VEGFR3/Flt4 axis regulates the growth and metastasis of breast tumors in an autocrine manner (13). The overexpression of VEGFR3 is also known to be associated with the prognosis of a variety of malignancies (14,15). The relationship between VEGFR3 and lymph node metastasis in gastric cancer has been reported. For example, Han et al. (16) confirmed that the expression of VEGFC and VEGFR3 in gastric cancer is related to lymph node metastasis. Choi et al. (17) found that VEGFR3-positive lymphatic density is significantly increased in gastric cancer and correlates with the primary tumor size, lymphatic invasion, and lymph node metastasis. The results of our experiment are consistent with previous studies confirming that VEGFR3 protein expression is related to tumor invasion depth and lymph node metastasis, but not to patients’ gender, age, tumor size, general typing and degree of differentiation (18,19), suggesting that VEGFR3 is involved in the occurrence and development of EGC and is correlated with tumor invasion and metastasis. In addition, studies on tumor biotherapy have shown that blocking the VEGFR3-mediated signal transduction pathway can effectively inhibit tumor lymphangiogenesis (20), suggesting that VEGFR3 is also a promising molecular target for aggressive EGC.
DNA methylation is a biochemical modification process catalyzed by DNA methyltransferase, which transfers methyl to a specific base with s-adenosine methionitrate as a methyl donor. In mammals, DNA methylation mainly occurs on the 5-carbon atom of the pyrimidine ring of cytidine nucleoside, and forms CpG with its 3-terminal guanine, appearing in double-chain symmetry. These pieces of DNA-rich CpG sequences are called CpG islands (21,22). Abnormal DNA methylation is one of the common mechanisms of inactivation or activation of key factors leading to cancer. For example, through the detection of methylation-related genes in the process of colorectal cancer carcinogenesis, researchers have found that the hypermethylation of eight genes is related to a reduction of transcription level, whereas the hypomethylation of three genes is related to an increase in transcription level (23). In recent years, through analysis of the cancer genome, gene expression profile, and gene methylation profile, an increasing number of differentially methylated genes related to tumor development and prognosis have been found (24-27). Therefore, the detection of gene methylation can be an effective biomarker for the early diagnosis and prognosis of various tumors including gastric cancer, central nervous system tumor, lung adenocarcinoma, uterine tumor, thyroid papillary carcinoma, and even targeted therapy (5,28-31).
Hypermethylation of tumor suppressor gene promoters and hypomethylation of global DNA are common in cancer tissues. In the VEGFR3 promoter, proximal GC-rich elements interact with Sp1 and Sp3 to activate transcription, and these CpG islands are also associated with the epigenetic silencing of gene expression in many cancer cell lines (32,33). For instance, the methylation status of three VEGFR gene promoters in 128 head and neck squamous cell carcinoma patients and their correlation with clinical characteristics were systematically examined in a previous study, and it was found that methylation of the VEGFR3 promoter was associated with poor prognosis (34). In this study, the methylation status of VEGFR3 gene promoter in EGC tissues and normal tissues was detected by MSP, suggesting that hypomethylation of VEGFR3 gene promoter is one of the main reasons for the upregulated expression of this gene in the development of EGC. Further analysis of the correlation between the methylation status of VEGFR3 gene promoter and the clinicopathological factors of EGC revealed that the frequency of methylation of VEGFR3 gene promoter was significantly correlated with the presence of lymph node metastasis (P<0.05). Therefore, detection of the methylation status of the VEGFR3 gene promoter is expected to be one of the biomarkers for the molecular diagnosis and disease stage assessment of this malignant disease.
Conclusions
In summary, our results indicate that VEGFR3 is closely related to the occurrence and development of EGC, and promoter methylation is one of the major mechanisms underlying inhibition of VEGFR3 overexpression in normal cells. To validate the methylation biomarkers for diagnosis and prognosis, clinical studies with larger sample sizes are needed that use different methylation detection methods such as Sanger sequencing and deep sequencing, to optimize the accuracy and reliability of the laboratory results. At present, several genes with promising experimental results are expected to be applicable for the clinical diagnosis and treatment of gastric cancer (35-39). Due to the continually evolving technologies for detecting methylation, detection of DNA methylation may be clinically applied in the future.
Acknowledgments
We would like to thank the Department of Pathology of Qilu Hospital (Shandong University) for providing case information, and Min Yang from the Department of Pathology of Qilu Hospital (Shandong University) for helping conducting the experiments. We would also like to thank the teachers who helped me with the experiment in the school of medicine of Shandong University. This manuscript has been edited and proofread by Medjaden Bioscience Limited.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.74). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Regional Ethics Committee of Qilu Hospital of Shandong University [No. KYLL-2017(KS)-399]. Because this was a retrospective study based on the medical records and tumor slides assessment, the written informed consent requirement was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fitzmaurice C, Dicker D, Pain A, et al. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505-27. [Crossref] [PubMed]
- Wang J, Taylor A, Showeil R, et al. Expression profiling and significance of VEGF-A, VEGFR2, VEGFR3 and related proteins in endometrial carcinoma. Cytokine 2014;68:94-100. [Crossref] [PubMed]
- de Aquino AR, Nonaka CF, de Carvalho CH, et al. Immunoexpression of VEGFR-3, but not the immunoexpression of VEGF-C or lymphatic density, is correlated with metastasis in lower lip squamous cell carcinoma. Int J Oral Maxillofac Surg 2017;46:16-23. [Crossref] [PubMed]
- Li D, Zhang L, Liu Y, et al. Specific DNA methylation markers in the diagnosis and prognosis of esophageal cancer. Aging (Albany NY) 2019;11:11640-58. [Crossref] [PubMed]
- Wang R, Zhu H, Yang M, et al. DNA methylation profiling analysis identifies a DNA methylation signature for predicting prognosis and recurrence of lung adenocarcinoma. Oncol Lett 2019;18:5831-42. [PubMed]
- Xue J, Gao HX, Sang W, et al. Identification of core differentially methylated genes in glioma. Oncol Lett 2019;18:6033-45. [PubMed]
- Klutstein M, Moss J, Kaplan T, et al. Contribution of epigenetic mechanisms to variation in cancer risk among tissues. Proc Natl Acad Sci U S A 2017;114:2230-4. [Crossref] [PubMed]
- Hamilton SR, Aaltonen LA. Translated by Yu JY. Pathology and genetics of digestive system tumors. Beijing: People's Health Publishing House 2006:355-75.
- Shibuya M. Tyrosine Kinase Receptor Flt/VEGFR Family: Its Characterization Related to Angiogenesis and Cancer. Genes Cancer 2010;1:1119-23. [Crossref] [PubMed]
- Xu HM, Zhu JG, Gu L, et al. VEGFR2 Expression in Head and Neck Squamous Cell Carcinoma Cancer Cells Mediates Proliferation and Invasion. Asian Pac J Cancer Prev 2016;17:2217-21. [Crossref] [PubMed]
- Zhou Z, Zhao C, Wang L, et al. A VEGFR1 antagonistic peptide inhibits tumor growth and metastasis through VEGFR1-PI3K-AKT signaling pathway inhibition. Am J Cancer Res 2015;5:3149-61. [PubMed]
- Zhao L, Zhu Z, Yao C, et al. VEGFC/VEGFR3 Signaling Regulates Mouse Spermatogonial Cell Proliferation via the Activation of AKT/MAPK and Cyclin D1 Pathway and Mediates the Apoptosis by affecting Caspase 3/9 and Bcl-2. Cell Cycle 2018;17:225-39. [Crossref] [PubMed]
- Varney ML, Singh RK. VEGF-C-VEGFR3/Flt4 axis regulates mammary tumor growth and metastasis in an autocrine manner. Am J Cancer Res 2015;5:616-28. [PubMed]
- Sha M, Jeong S, Chen XS, et al. Expression of VEGFR-3 in intrahepatic cholangiocarcinoma correlates with unfavorable prognosis through lymphangiogenesis. Int J Biol Sci 2018;14:1333-42. [Crossref] [PubMed]
- Chen B, Liu J, Wang X, et al. Co-expression of PDGF-B and VEGFR-3 strongly correlates with poor prognosis in hepatocellular carcinoma patients after hepatectomy. Clin Res Hepatol Gastroenterol 2018;42:126-33. [Crossref] [PubMed]
- Han FH, Li HM, Zheng DH, et al. The effect of the expression of vascular endothelial growth factor (VEGF)-C and VEGF receptor-3 on the clinical outcome in patients with gastric carcinoma. Eur J Surg Oncol 2010;36:1172-9. [Crossref] [PubMed]
- Choi JH, Oh YH, Park YW, et al. Correlation of vascular endothelial growth factor-D expression and VEGFR-3-positive vessel density with lymph node metastasis in gastric carcinoma. J Korean Med Sci 2008;23:592-7. [Crossref] [PubMed]
- Li XF, Zhang YX, Zhang TG, et al. The Expression of VEGFR2 and VEGFR3 in Early Gastric Cancer and Its Clinical Significance. J Bioprocess Biotech 2018;8:3. [Crossref]
- Ge H, Yan Y, Guo L, et al. Prognostic and clinical significance of VEGFR-3 in gastric cancer: A meta-analysis. Clin Chim Acta 2017;474:114-9. [Crossref] [PubMed]
- Chaudary N, Milosevic M, Hill RP. Suppression of vascular endothelial growth factor receptor 3 (VEGFR3) and vascular endothelial growth factor C (VEGFC) inhibits hypoxia-induced lymph node metastases in cervix cancer. Gynecol Oncol 2011;123:393-400. [Crossref] [PubMed]
- Dmitrijeva M, Ossowski S, Serrano L, et al. Tissue-specific DNA methylation loss during ageing and carcinogenesis is linked to chromosome structure, replication timing and cell division rates. Nucleic Acids Res 2018;46:7022-39. [Crossref] [PubMed]
- Edwards JR, Yarychkivska O, Boulard M, et al. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017;10:23. [Crossref] [PubMed]
- Sun X, Chen D, Jin Z, et al. Genome-wide methylation and expression profiling identify methylation-associated genes in colorectal cancer. Epigenomics 2020;12:19-36. [Crossref] [PubMed]
- Liu X, Fu J, Bi H, et al. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer 2019;19:1212. [Crossref] [PubMed]
- Wei S, Yun X, Ruan X, et al. Identification of potential pathogenic candidates or diagnostic biomarkers in papillary thyroid carcinoma using expression and methylation profiles. Oncol Lett 2019;18:6670-8. [PubMed]
- Pickles JC, Fairchild AR, Stone TJ, et al. DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: a population-based study. Lancet Child Adolesc Health 2020;4:121-30. [Crossref] [PubMed]
- Kishibuchi R, Kondo K, Soejima S, et al. DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common epigenetic alteration in thymic carcinoma. Int J Oncol 2020;56:315-26. [PubMed]
- Tahara T, Arisawa T. DNA methylation as a molecular biomarker in gastric cancer. Epigenomics 2015;7:475-86. [Crossref] [PubMed]
- Karimi S, Zuccato JA, Mamatjan Y, et al. The central nervous system tumor methylation classifier changes neuro-oncology practice for challenging brain tumor diagnoses and directly impacts patient care. Clin Epigenetics 2019;11:185. [Crossref] [PubMed]
- Kommoss FKF, Stichel D, Schrimpf D, et al. DNA methylation-based profiling of uterine neoplasms: a novel tool to improve gynecologic cancer diagnostics. J Cancer Res Clin Oncol 2020;146:97-104. [Crossref] [PubMed]
- Park JL, Jeon S, Seo EH, et al. Comprehensive DNA Methylation Profiling Identifies Novel Diagnostic Biomarkers for Thyroid Cancer. Thyroid 2020;30:192-203. [Crossref] [PubMed]
- Quentmeier H, Eberth S, Romani J, et al. DNA methylation regulates expression of VEGF-R2 (KDR) and VEGF-R3 (FLT4). BMC Cancer 2012;12:19. [Crossref] [PubMed]
- Hertel J, Hirche C, Wissmann C, et al. Transcription of the vascular endothelial growth factor receptor-3 (VEGFR3) gene is regulated by the zinc finger proteins Sp1 and Sp3 and is under epigenetic control: transcription of vascular endothelial growth factor receptor 3. Cell Oncol (Dordr) 2014;37:131-45. [Crossref] [PubMed]
- Misawa Y, Misawa K, Kawasaki H, et al. Evaluation of epigenetic inactivation of vascular endothelial growth factor receptors in head and neck squamous cell carcinoma. Tumour Biol 2017;39:1010428317711657. [Crossref] [PubMed]
- Zhang Z, Yu W, Chen S, et al. Methylation of the claudin3 promoter predicts the prognosis of advanced gastric adenocarcinoma. Oncol Rep 2018;40:49-60. [PubMed]
- Sui BQ, Zhang CD, Liu JC, et al. HOXB13 expression and promoter methylation as a candidate biomarker in gastric cancer. Oncol Lett 2018;15:8833-40. [PubMed]
- Deng P, Chang XJ, Gao ZM, et al. Downregulation and DNA methylation of ECRG4 in gastric cancer. Onco Targets Ther 2018;11:4019-28. [Crossref] [PubMed]
- Dai D, Zhou B, Xu W, et al. CHFR Promoter Hypermethylation Is Associated with Gastric Cancer and Plays a Protective Role in Gastric Cancer Process. J Cancer 2019;10:949-56. [Crossref] [PubMed]
- Harada H, Hosoda K, Moriya H, et al. Cancer-specific promoter DNA methylation of Cysteine dioxygenase type 1 (CDO1) gene as an important prognostic biomarker of gastric cancer. PLoS One 2019;14:e0214872. [Crossref] [PubMed]


