
Cyclophilin B overexpression predicts a poor prognosis and activates metastatic pathways in colon cancer
Introduction
Colorectal cancer is a leading cause of cancer-related death. The incidence of colorectal cancer ranks fourth among all malignant tumors, with approximately 140,000 new cases and 50,000 cases of death each year in the United States (1). In China, due to the huge population base and dramatic changes in the environment, the number of deaths per year is approximately 190,000 (2).
Colonoscopy has been used in the clinic for the early diagnosis of colorectal cancer and has promoted a 5-year survival rate of almost 90%. Unfortunately, many patients lose the chance for early diagnosis and effective treatment and often develop distant metastases, and the 5-year survival rate for those patients is only 12.5% (3). For these distant-stage patients, we more urgently need to find effective biomarkers closely related to prognosis and their pathological mechanisms in order for more precise targeted treatments.
Cyclophilins (Cyps) have been reported to exhibit peptidyl-prolyl isomerase enzymatic activity and are involved in a variety of cell functions (4,5). CypB (cyclophilin B) is a member of the Cyps family, which is predominantly located in the endoplasmic reticulum (ER) and was indicated to act as the target of cyclosporin A (an immunosuppressive drug). CypB has also been shown to be involved in many biological processes, including protein folding (6), virus replication (7), immunosuppression (8) and osteogenesis (9). Recently, a high level of CypB was found in pancreatic, breast, gastric and liver cancer (10-13). CypB was found to promote cancer by accelerating cell proliferation, decreasing cell apoptosis, and facilitating cell migration and invasion (10,14-16). However, the clinical significance of CypB overexpression remains to be investigated in colorectal cancer.
In this study, we analyzed the expression of CypB by RNAscope in situ hybridization and immunohistochemical (IHC) staining in colon cancer. Furthermore, we analyzed the correlation between CypB expression and clinicopathological characteristics. Then, we focused on the prognostic significance and signaling pathways of CypB in colon cancer. Our study demonstrates that CypB was overexpressed in colon cancer tissues and that the upregulation of CypB was associated with poor survival. Bioinformatics analysis and the in vitro study revealed that CypB was involved in tumor metastatic signaling pathways. Hence, we propose that CypB serves as a promising prognostic biomarker and may promote metastasis in colon cancer.
Methods
Patients and tumor tissue microarray (TMA)
The colonic TMA (HCol-Ade180Sur-07, Shanghai Outdo Biotech Co., Shanghai, China) used in RNAscope analysis contained 90 cases of colonic adenocarcinoma and paired adjacent noncancerous tissues. All tissues were retrospectively collected from patients after surgery from January 2009 to October 2009. Before surgery the patients did not receive any chemotherapy or radiotherapy. And the follow-up data of patients were acquired from February 2009 to May 2014. The included patients were followed-up routinely either till their expiry or at least 5 years from their surgery date. Detailed clinicopathological characteristics are listed in supplementary Table S1. The HCol-Ade030PG-01 TMA (Shanghai OUTDO Biotech Co., Shanghai, China) used in IHC analysis consisted of 15 paired colorectal adenocarcinoma tissues and matched normal mucosa; All tissues were retrospectively collected from patients underwent surgery from January 2009 to October 2009. Before surgery the patients did not receive any chemotherapy or radiotherapy. The TMAs were stored in 4 °C before use. This TMA has no clinicopathological or follow-up data. Tumor T staging, N staging and TNM staging were performed based on the 7th Edition of American Joint Committee on Cancer (AJCC) staging system. Histological grading was performed according to the World Health Organization (WHO) classification of tumors of the digestive system of 2010. According to the location of the tumor, tumors located before the splenic flexure of the transverse colon were defined as right colon tumors, and tumors located at or after the splenic flexure of the transverse colon were defined as left colon tumors. Our study design, tissue sample, and data collection were accomplished according to our institutional protocols, which approved by Institutional Ethics Committee, Beijing Chao-Yang Hospital of Capital Medical University (No. 2018-Research-61) and informed consent was taken from all the patients. Our primary endpoint of the study was overall survival (OS) that is stated as the time from the date of surgery to death or the last follow-up date.
RNAscope in situ hybridization and image analysis
RNAscope in situ hybridization analysis was performed on colon cancer TMAs using a probe that targeted human CypB (Cat. No. 476701; Advanced Cell Diagnostics, Hayward, CA, USA) based on the manufacturer’s instruction, and a standard pretreatment protocol was used. RNAscope 2.5 High Definition (HD) Reagent Kit-brown (Cat. No. 322310; Advanced Cell Diagnostics, Hayward, CA, USA) was adopted to amplify and visualize the hybridization signals. Then, the slide image was taken with an Aperio scanner and viewed with AperioImageScope software (v12.3.1.6002, Leica Biosystems). CypB mRNA molecules are shown as brown spots and were counted manually. According to the manufacturer’s guidelines, a 5-tier scoring system was developed for semiquantitative microscopic evaluations: score 0 (−), no staining or less than 1 dot in each of ten cells; score 1 (+), 1–3 dots per cell; score 2 (++), 4–10 dots per cell, very few dot clusters; score 3 (+++), >10 dots per cell and the cells with dot clusters were <10% of all cells; and score 4 (++++), >10 dots per cell and the cells with dot clusters were >10% of all cells. Scores of 0–2 were considered low CypB mRNA expression, and scores of 3–4 were considered high CypB mRNA expression. Bacillus subtilis DapB mRNA (Cat. No. 310043; Advanced Cell Diagnostics, Hayward, CA, USA) was probed as a negative control. All the staining scores were reviewed by two pathologists through blinded-reading.
IHC staining analysis
The TMA slide was deparaffinized and rehydrated and rinsed in water. To quench endogenous peroxidase activity, the TMA slide was treated with 0.3% H2O2 for 10 minutes at room temperature. Antigen retrieval was performed in 0.01 M sodium citrate (pH =6.0) with heating in a pressure cooker. The sections were then blocked in 2% goat serum and were incubated with the primary antibody for 1 hour at room temperature. This study used rabbit polyclonal anti-CypB antibody (ab16045, Abcam Inc., Cambridge, MA, USA) as the primary antibody with 1:500 dilution. Then the second antibody from SP reagent kit (Zhongshan Goldenbridge Biotechnology Co., Beijing, China) was exerted to incubate the TMA sections for 20 minutes at room temperature, followed by further incubation with streptavidin-horseradish peroxidase complex. Staining with 3,3'-diaminobenzidine kit (DAB; Zhongshan Goldenbridge Biotechnology Co.), TMA sections were counter-stained with hematoxylin and evaluated. Score is the combination of staining intensity (0= negative, 1= mild staining, 2= moderate staining and 3= strong staining) and percentage of positive cells (0: <5%, 1: 6% to 25%, 2: 26% to 50%, 3: 51% to 75% and 4: >76%) (17). Finally the CypB staining was assigned to one of 4 levels as follows: negative (−) (score of 0), weak (+) (score of 1–4), moderate (++) (score of 5–8) to strong (+++) (score of 9–12). Negative (−) and weak (+) were considered as low expression, and moderate (++) and strong (+++) were considered as high expression.
Colon Adenocarcinoma (COAD) RNA-seq data from the Cancer Genome Atlas (TCGA)
The COAD RNA-seq datasets of TCGA, which enrolled 286 COAD tissues and 41 adjacent noncancerous tissues, were downloaded through the UCSC cancer genome browser (https://xenabrowser.net). The Illumina HiSeq 2000 RNA Sequencing platform was used to experimentally measure gene expression at the University of North Carolina TCGA genome characterization center. Level 3 data was downloaded from TCGA data coordination center. This dataset shows the gene-level transcription estimates, as in log2(x+1) transformed RSEM normalized count.
Gene set enrichment analysis (GSEA) and network construction
Gene Set Enrichment Analysis (GSEA, http://software.broadinstitute.org/gsea/) was applied for enriching CypB related pathways. At first, the top 50 up-regulated and 50 down-regulated differential genes between normal and cancer tissues from COAD datasets of TCGA were selected using Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) (18). Finally, 87 genes were selected after deleting non-coding RNA. Then CypB related signaling pathways were enriched using GSEA by dividing those differential genes into two sets according to the median value of CypB. The gene set permutations analysis was repeated 1,000 times, according to the default weighted enrichment statistical method. Nominal P value, enrichment score (ES) and false discovery rate (FDR) were calculated to verify the significant difference for GSEA. After gene enrichment, the Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) was used to construct protein-protein interactions (PPI) and screen the CypB related signaling pathways.
Cell lines, cell culture and cell transfection
The human colon cancer cell line HCT116 purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA) was used for our experiment. Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; HyClone; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37 °C in a humidified atmosphere containing 5% CO2. HCT116 cells were seeded in six-well plates and allowed to attach overnight. With the application of lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), CypB small interfering RNA (siRNA) and control siRNA were transfected into the cells respectively according to the manufacturer’s recommendations. Then the cells were further cultured at 37 °C in a 5% CO2 atmosphere. CypB siRNA-1 sequence was 5'-GCAUGGAGGUGGUGCGG-3', CypB siRNA-2 sequence was 5'-CUUAGCUACAGGAGAGAA-3', and the negative control siRNA sequence was 5'-TTCTCCGAACGTGTCACGT-3'. Both of them were designed and synthesized by the Beijing Hesheng Gene Technology Co., Ltd. (Beijing, China).
RNA extraction and real-time quantitative PCR
Total cellular RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was then reverse transcribed to cDNA using the EasyScript® First-Strand cDNA Synthesis kit (Transgene, Beijing, China). Gene expression analysis was performed by qRT-PCR using a SYBR Premix Ex Taq Kit (Takara, Dalian, China). Relative gene expression was quantified using the comparative threshold cycle (2−ΔΔCt) method. The PCR program was as follows: pre-denaturation at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 5 s, and annealing and elongation at 60 °C for 30 s. The primers used in the experiment were as follows:
CypB: Forward, AAGTCACCGTCAAGGTGTATTTT; Reverse, TGCTGTTTTTGTAGCCAAATCCT.
CNN1: Forward, AGGTTAAGAACAAGCTGGCCC; Reverse, ATGAAGTTGTTGCCGATGCG.
MYL9: Forward, CTCGCTGGGGAAGAACCCC; Reverse, CGTTGCGAATCACATCCTCG.
MYH11: Forward, AGACACAAGTATCACGGGAGAG; Reverse, TTGCCGAATCGTGAGGAGTT.
E-cadherin: Forward, GTCACTGACACCAACGATAATCCT; Reverse, TTTCAGTGTGGTGATTACGACGTTA.
Snail: Forward, GCCATGTCCGGACCCACACTG; Reverse, GGCAGGGGCAGGTATGGAGA.
TWIST: Forward, GTCCGCAGTCTTACGAGGAG; Reverse, GCTTGAGGGTCTGAATCTTGCT.
Vimentin: Forward, CCTGAACCTGAGGGAAACTAA; Reverse, GCAGAAAGGCACTTGAAAGC.
18s: Forward, AAACGGCTACCACATCCA; Reverse, CACCAGACTTGCCCTCCA.
Statistical analysis
Statistical analyses were conducted using SPSS software for Windows, version 17.0 (SPSS, Chicago, IL, USA). GraphPad Prism for Windows, version 5.0 (GraphPad Software, San Diego, CA, USA) was used to create the artwork. Quantitative variables were compared by means of the student t-test. Categorical variables were compared using the χ2 test. The Cox proportional hazards regression model and the Kaplan-Meier test were used to assess the OS rates. The survival curves were plotted by the log-rank test. P<0.05 was considered statistically significant.
Results
RNAscope in situ hybridization and IHC staining present the overexpression of CypB in colon cancer
TMA that contained 90 paired cancer and adjacent normal tissues was used to determine the expression of CypB. Finally, 80 cancer tissues and 84 adjacent normal tissues were successfully stained to show the mRNA levels of CypB by RNAscope. According to the expression level of CypB mRNA (representative images were provided in Figure 1), staining intensities of score 0 (−), score 1 (+) and score 2 (++) were classified as the low expression group, and score 3 (+++) and score 4 (++++) were classified as the high expression group. CypB mRNA was found to be significantly overexpressed in colon cancer tissues compared with adjacent normal tissues (P<0.001; Table 1).
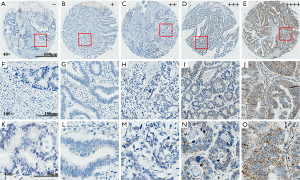
Table 1
| Histological type | Case numbers | CypB expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Tumor tissues | 80 | 39 | 41 | <0.001* |
| Nontumor tissues | 84 | 70 | 14 | |
*, P value less than 0.05. CypB, cyclophilin B.
We also used TMA with a small sample size to detect the expression of CypB protein (Figure 2). As in mRNA level, the expression of CypB protein in colon cancer is significantly higher than that in adjacent tissues (P<0.05; Table 2).
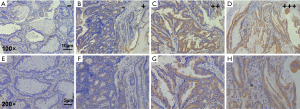
Table 2
| Histological type | Case numbers | CypB expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Tumor tissues | 15 | 10 | 5 | 0.042* |
| Nontumor tissues | 15 | 15 | 0 | |
*, P value less than 0.05. CypB, cyclophilin B.
Clinicopathological analysis reveals that CypB is associated with advanced T stage
The correlation between CypB levels and the clinicopathological parameters of 80 colon cancer patients was analyzed. The clinicopathological data of the patients were summarized in the supplementary Table S1. Our analysis indicated that the levels of CypB were significantly higher in patients with T4 stage than in those with T1–3 stage (P=0.043; Table 3). However, there were no significant correlations between the levels of CypB and other parameters, including age, sex, tumor size, N stage, histological grade, TNM stage and tumor position (P>0.05; Table 3).
Table 3
| Parameters | Group | Case numbers | CypB expression | P value | |
|---|---|---|---|---|---|
| Low expression | High expression | ||||
| Age | <65 | 33 | 18 | 15 | 0.385 |
| ≥65 | 47 | 21 | 26 | ||
| Sex | Male | 43 | 21 | 22 | 0.987 |
| Female | 37 | 18 | 19 | ||
| Tumor size | <5 cm | 33 | 14 | 19 | 0.296 |
| ≥5 cm | 46 | 25 | 21 | ||
| NA | 1 | 0 | 1 | ||
| T stage | T1–3 | 55 | 31 | 24 | 0.043* |
| T4 | 25 | 8 | 17 | ||
| N stage | N0 | 53 | 28 | 25 | 0.306 |
| N1–2 | 27 | 11 | 16 | ||
| Histological grade | I–II | 69 | 34 | 35 | 0.814 |
| III–V | 11 | 5 | 6 | ||
| TNM stage | I–II | 51 | 26 | 25 | 0.597 |
| III–V | 29 | 13 | 16 | ||
| Tumor position | Left colon | 37 | 15 | 22 | 0.17 |
| Right colon | 41 | 23 | 18 | ||
| NA | 2 | 1 | 1 | ||
*, P value less than 0.05. CypB, cyclophilin B; TNM, tumor-node-metastasis.
CypB mRNA overexpression predicts a poor prognosis of colon cancer patients
The prognostic significance of CypB mRNA expression was further investigated in colon cancer patients. In total, 80 patients were followed up for 0.4–64 months (mean ± SD, 47.02±19.48 months). At the end of follow up, 27 patients had died. Kaplan-Meier analysis revealed that the high expression of CypB was associated with a shorter OS (Figure 3A, P=0.0139). Univariate and multivariate Cox regression analyses indicated that TNM stage (P=0.000) and CypB expression (P=0.015) were independent prognostic indicators for poor survival (Table 4). Furthermore, subgroup analysis indicated that high levels of CypB were associated with poor survival for patients with stage T3–4, lymph node metastasis, tumor size ≥5 cm or right colonic cancer (Figure 3B,C,D,E, P<0.05). In addition, our analysis also indicated that patients in TNM stages III–IV with high CypB expression had a shorter survival time, although the difference was not significant (Figure 3F, P=0.0616).
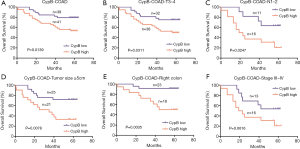
Table 4
| Parameters | OS | ||||||
|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (<65 vs. ≥65) | 1.067 | 0.495–2.301 | 0.868 | ||||
| Sex (male vs. female) | 1.832 | 0.822–4.080 | 0.138 | ||||
| Tumor size (<5 vs. ≥5 cm) | 3.183 | 1.282–7.902 | 0.013* | ||||
| T stage (T1–3 vs. T4) | 3.432 | 1.599–7.364 | 0.002* | ||||
| N stage (N0 vs. N1–2) | 3.702 | 1.712–8.006 | 0.001* | ||||
| Histological grade (I–II vs. III–IV) | 3.534 | 1.485–8.411 | 0.004 | ||||
| TNM stage (I–II vs. III–IV) | 4.805 | 2.146–10.755 | 0.000* | 4.918 | 2.193–11.03 | 0.000* | |
| Tumor position (left vs. right colon) | 0.586 | 0.269–1.277 | 0.179 | ||||
| CypB mRNA expression (low vs. high) | 2.693 | 1.178–6.155 | 0.019* | 0.36 | 0.157–0.823 | 0.015* | |
*, P value less than 0.05. CI, confidence interval; CypB, cyclophilin B; HR, hazard ratio; OS, overall survival; TNM, tumor-node-metastasis.
Validation of CypB overexpression and its prognostic significance in COAD RNA-seq dataset of TCGA
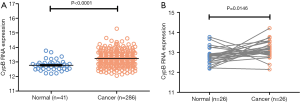
To further validate our findings, we analyzed the CypB mRNA expressions in COAD RNA-seq dataset of TCGA. First, we compared the CypB mRNA levels between 286 cancerous tissues and 41 normal tissues (Figure 4A). As expected, the CypB levels were significantly higher in cancer tissues than in normal tissues (P<0.0001). Furthermore, in the TCGA 26 paired cancer and corresponding normal tissues, the CypB mRNA levels were also markedly increased in cancer tissues compared to normal tissues (Figure 4B, P=0.0146).
Next, we determined the prognostic significance of CypB mRNA in 286 COAD patients. The CypB mRNA expression levels and clinicopathological parameters are summarized in the supplementary Table S2. OS differences between patients with high or low CypB expression were analyzed by Cox regression models and log-rank tests. As shown by Kaplan-Meier plots, a high level of CypB mRNA was associated with a reduced OS time (P=0.048, Figure 5A). In subgroup analysis, we found that a higher level of CypB mRNA was associated with a shorter OS time for patients with advanced tumors, such as in patients with stage T3–4, lymph node metastasis and TNM stage III-IV (Figure 5B,C,D, P<0.05). Furthermore, Cox multivariate analyses confirmed that CypB mRNA was associated with the OS time of COAD patients (Table 5, P=0.007).
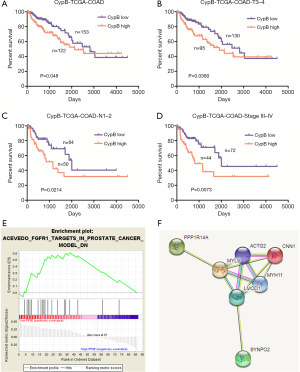
Table 5
| Parameters | OS | ||||||
|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (<65 vs. ≥65) | 1.521 | 0.914–2.531 | 0.106 | ||||
| Sex (male vs. female) | 0.686 | 0.420–1.121 | 0.133 | ||||
| T stage (Tis–2 vs. T3–4) | 2.516 | 1.009–6.271 | 0.048* | ||||
| N stage (N0 vs. N1–2) | 2.432 | 1.499–3.947 | 0.000* | ||||
| TNM stage (I–II vs. III–IV) | 2.548 | 1.538–4.219 | 0.000* | 2.808 | 1.688–4.671 | 0.000* | |
| Tumor position (left vs. right colon) | 1.325 | 0.779–2.254 | 0.299 | ||||
| CypB mRNA expression (low vs. high) | 1.614 | 0.998–2.612 | 0.049* | 2.007 | 1.212–3.323 | 0.007* | |
*, P value less than 0.05. CI, confidence interval; CypB, cyclophilin B; HR, hazard ratio; OS, overall survival; TNM, tumor-node-metastasis.
GSEA and STRING analyses indicate that CypB is enriched in the metastatic pathways
To identify potential function of CypB, we performed GSEA using TCGA data. The cut-off criterion is set to nominal P value <0.05 and |enrichment score (ES)| >0.55. As shown in Figure 5, the gene set “FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_DN” was enriched with CypB lowly expressed (Figure 5E, P<0.05). This gene set is involved in the regulation of epithelial-to-mesenchymal transition (EMT) and Wnt signaling pathway (19). Furthermore, the enriched genes were analyzed by STRING to generate visual images of PPIs and the potential biological processes (Figure 5F). The results uncovered that CypB was closely involved in tumor metastatic pathways, including cell adhesion, tight junction, cell-cell junction organization, extracellular matrix organization and adherens junctions interactions (Table S3).
CypB is associated with myosin related genes and may involve in Snail-mediated EMT in colon cancer
Based on the enriched gene set with GSEA analysis, we found that the expressions of calponin 1 (CNN1), myosin light chain 9 (MYL9) and myosin heavy chain 11 (MYH11) were positively correlated with the expression of CypB. These three genes are all necessary in cell movement, cytokinesis and spindle formation, which are related to tumor invasion and metastasis. Therefore, in vitro experiments by knockdown of CypB in colon cancer cell HCT116 were performed to verify the bioinformatics results. We found that compared with the NC-siRNA group, CypB silencing significantly reduced the expressions of MYL9, MYH11 and CNN1 (Figure 6A,B,C,D, P<0.05).
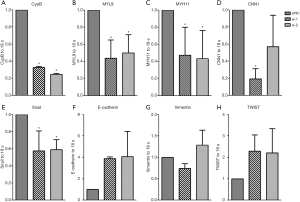
Subsequently, GSEA and STRING analyses revealed that CypB may closely involved in tumor metastatic pathways, such as EMT. During EMT, epithelial cells lose epithelial characteristics and acquire a mesenchymal, highly invasive phenotype. In this process, many transcriptional regulators, such as TWIST, ZEB, Snail and Slug are activated, leading to the downregulation of E-cadherin expression. In HCT116 cells, CypB silencing significantly reduced Snail expression (Figure 6E, P=0.0048). Although there was no statistical significance, the expression of E-cadherin increased (Figure 6F, P>0.05) after CypB decreased. But there were no significant changes in Vimentin and TWIST expressions (Figure 6G,H, P>0.05). These data suggest that Snail-mediated EMT may be associated with CypB in colon cancer.
Discussion
Previous studies have found that CypB was involved in many pathophysiological processes, including osteogenesis, hepatitis virus replication, and immunosuppression. In recent years, CypB overexpression has been observed in stomach, liver, pancreatic, breast and several other types of cancers (10,11,13,20,21). Some in vitro studies have shown that CypB could promote tumor cell proliferation, protect tumor cells against oxidative stress, and stimulate neovascularization (22-24). So far, only one research team has analyzed the relationship between CypB expression and prognosis in colon cancer. Their research was only at the protein level, and the correlation between CypB and clinicopathological parameters was not further explored (12). Our study applied a new technique of RNA in situ hybridization-RNAscope, and explored the expression pattern and clinical significance of CypB in colon cancer at the RNA level. We also detected the CypB protein expression using IHC. Bioinformatics analysis was applied to find out the CypB involved signaling pathways, which provides a new clue to reveal the function of CypB in colon cancer.
For formalin-fixed, paraffin-embedded tissue sections, immunohistochemistry remains the overwhelming technique of choice. However, validations can be complex, with significant specificity, sensitivity and reproducibility issues. Commercial antibodies from many available vendors may also lead to nonstandard approaches. The RNAscope in situ hybridization method enabled a realistic alternative with fewer validation steps and more stringent and reproducible assessment criteria (25,26). In our analyses, we used this method to stain CypB mRNA in single colon cancer cells and adjacent normal cells. We also analyzed the CypB protein expression using IHC. We found that CypB mRNA and protein were distributed in the cytoplasm and nucleus. Furthermore, we observed that CypB was apparently overexpressed in colon cancer tissues compared with adjacent normal tissues. The high expression of CypB mRNA was significantly higher in patients with T4 stage than in those with T1–3 stage. However, there were no significant correlations between CypB mRNA expression and other parameters. To the best of our knowledge, this is the first study to demonstrate the relation between CypB and clinicopathological parameters in colon cancer.
Additionally, the patients who had relatively high levels of CypB showed poorer prognoses than their low-level counterparts, and further Cox regression analyses indicated that CypB mRNA expression was an independent prognostic indicator. The expression of CypB is not significantly correlated with clinicopathological parameters, such as T and N stages, but its high expression is related to a poor prognosis, suggesting that CypB may not directly promote the tumor proliferation but may affect the prognosis in other ways. For example, Choi’s study found that the overexpression of CypB could promote oxaliplatin resistance and inhibit oxaliplatin-induced apoptosis in colon cancer cells (27), therefore, further research is needed on this perspective. In addition, in subgroup analysis, we found that CypB had prognostic significance in more advanced tumors, such as in patients with T3–4, lymph node metastasis and clinical stage III-IV, suggesting that CypB may play a vital role in late stage of colon cancer, such as promoting cancer migration.
With the wide application of sequencing technology, TCGA datasets contain differentially expressed transcripts of many cancers (28,29). Here, the COAD RNA-seq dataset in the TCGA was downloaded and analyzed. We confirmed that CypB mRNA was highly upregulated and served as a prognostic biomarker in colon cancer, especially in more advanced tumors. These results further validate our main findings from the TMA.
The mechanism and signaling pathways which CypB is involved in several cancers are studied in depth (10,15,21,30,31). However, the detailed mechanism for CypB in colon cancer progression still needs to be elucidated. In our study, bioinformatic analysis showed that the CypB may be closely associated with metastatic related processes, such as EMT and Wnt signaling pathway. Further cell experiments revealed that compared with the NC-siRNA group, CypB silencing significantly reduced the expressions of MYL9, MYH11 and CNN1. These genes all belong to the myosin family and more and more evidences show that this family may play important roles in tumor invasion and metastasis development, including EMT process (32,33). During EMT, epithelial cells lose epithelial characteristics and acquire a mesenchymal, highly invasive phenotype (34,35). Therefore we next tested several EMT related genes after knockdown of CypB. And results showed that CypB may be associated with Snail-mediated EMT in colon cancer. But further in vivo experiments should be designed to verify our findings in vitro.
Conclusions
Collectively, we report here that CypB is remarkedly overexpressed in human colon cancer. Overexpressed CypB is an independent prognostic indicator for poor survival, especially for advanced tumors. Bioinformatic and in vitro study analysis revealed that CypB is associated with some myosin related genes and may involve in Snail-mediated EMT process in colon cancer. CypB may have an important role in the regulation of tumor metastasis. In this regard, we suggest that CypB could serve as a promising poor prognostic biomarker for colon cancer.
Table S1
| Tissue code | CypB expression* | OS (m) | OS# | Age (y) | Sex& | T stage | N stage | M stage | TNM stage | Histological grade | Tumor position | Tumor size (cm3) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D15A1454-B30-C1 | 1 | 64 | 0 | 72 | 1 | T3 | N0 | M0 | 2A | II | Right colon | 6.5×5×1.7 |
| D15A1455-B30-C1 | 1 | 1 | 57 | 0 | T2 | N1b | M0 | 3A | II | Right colon | 6.5×5.5×2 | |
| D15A1456-B30-C1 | 1 | 13 | 1 | 76 | 1 | T3 | N0 | M0 | 2A | II | Descending colon | 5.5×4.5×2 |
| D15A1458-B30-C1 | 1 | 64 | 0 | 63 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 4.5×3.5×1.5 |
| D15A1516-B30-C1 | 76 | 0 | 53 | 1 | T4b | N0 | M0 | 2C | I–II | Sigmoid colon | 5×3×1.5 | |
| D15A1461-B30-C1 | 2 | 63 | 0 | 78 | 0 | T3 | N2b | M0 | 3C | II | Ascending colon | 7×5×1 |
| D15A1464-B30-C1 | 2 | 7 | 1 | 63 | 1 | T4a | N2b | M0 | 3C | III | Sigmoid colon | 5.5×4.5×1.5 |
| D15A1462-B30-C1 | 1 | 22 | 1 | 78 | 1 | T3 | N0 | M1b | 4B | I–II | Sigmoid colon | 7.5×3×1.5 |
| D15A1502-B30-C1 | 1 | 63 | 0 | 68 | 0 | T3 | N0 | M0 | 2A | II | Hepatic flexure | 6×3×2 |
| D15A1503-B30-C1 | 1 | 63 | 0 | 39 | 1 | T4a | N0 | M0 | 2B | II | Transverse colon | 6×4×3 |
| D15A1504-B30-C1 | 23 | 1 | 68 | 1 | T2 | N0 | M0 | 1 | I–II | Hepatic flexure | 5.5×4×2 | |
| D15A1505-B30-C1 | 1 | 63 | 0 | 62 | 1 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 2.5×2×0.5 |
| D15A1508-B30-C1 | 1 | 63 | 0 | 78 | 1 | T3 | N0 | M0 | 2A | II | Ascending colon | 5×4×2 |
| D15A1510-B30-C1 | 2 | 44 | 1 | 50 | 0 | T4a | N0 | M0 | 2B | II | Hepatic flexure | 4×3.5×1 |
| D15A1556-B30-C1 | 2 | 62 | 0 | 73 | 1 | T3 | N0 | M0 | 2A | I–II | Hepatic flexure | 11×6×2 |
| D15A1557-B30-C1 | 2 | 38 | 1 | 68 | 1 | T4a | N0 | M0 | 2B | II | Hepatic flexure | 6×4×1 |
| D15A1558-B30-C1 | 2 | 13 | 1 | 87 | 0 | T4b | N1b | M0 | 3C | I–II | Right colon | 6×4×1 |
| D15A1559-B30-C1 | 2 | 8 | 1 | 52 | 0 | T4a | N0 | M0 | 2B | II–III | Sigmoid colon | 7×5×2.5 |
| D15A1560-B30-C1 | 2 | 62 | 0 | 51 | 0 | T1 | N0 | M0 | 1 | II | Sigmoid colon | 2.7×1.7×1.3 |
| D15A1561-B30-C1 | 2 | 56 | 1 | 55 | 1 | T4a | N2a | M0 | 3C | II | Splenic flexure | 3.5×3.5×1 |
| D15A1562-B30-C1 | 1 | 17 | 1 | 73 | 1 | T4a | N0 | M1b | 4B | III–IV | Ascending colon | 6.5×5×1.5 |
| D15A1563-B30-C1 | 1 | 62 | 0 | 61 | 0 | T3 | N0 | M0 | 2A | II | Right colon | 3×3×2 |
| D15A1564-B30-C1 | 12 | 1 | 48 | 0 | T3 | N0 | M0 | 2A | II–III | Transverse colon | 1.5×1×1 | |
| D15A1565-B30-C1 | 1 | 62 | 0 | 59 | 0 | T2 | N0 | M0 | 1 | II | Sigmoid colon | 3×2.5×1 |
| D15A1566-B30-C1 | 40 | 1 | 77 | 0 | T2 | N0 | M0 | 1 | II | Sigmoid colon | 4×4×3 | |
| D15A1567-B30-C1 | 2 | 42 | 1 | 78 | 1 | T3 | N1a | M0 | 3B | II | Ascending colon | 5×5×1.5 |
| D15A1570-B30-C1 | 1 | 62 | 0 | 31 | 1 | T3 | N1b | M0 | 3B | I–III | Ascending colon | 4×3×1 |
| D15A1571-B30-C1 | 33 | 1 | 79 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 7×5×2 | |
| D15A1572-B30-P1 | 1 | 61 | 0 | 81 | 1 | T3 | N1b | M0 | 3B | II | Sigmoid colon | 4×3×1 |
| D15A1573-B30-C1 | 2 | 61 | 0 | 85 | 1 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 4.3×2×0.5 |
| D15A1574-B30-C1 | 2 | 40 | 1 | 90 | 1 | T4a | N0 | M0 | 2B | II | Sigmoid colon | 7×5×5 |
| D15A1576-B30-C1 | 1 | 61 | 0 | 70 | 0 | T2 | N0 | M0 | 1 | II | Sigmoid colon | 4.5×2×1 |
| D15A1577-B30-C1 | 2 | 23 | 1 | 66 | 0 | T4b | N1b | M0 | 3C | II–III | Ascending colon | 5×4×1.5 |
| D15A1579-B30-P1 | 2 | 61 | 0 | 73 | 1 | T3 | N0 | M0 | 2A | II | Descending colon | 3.5×3×1 |
| D15A1614-B30-C1 | 2 | 61 | 0 | 54 | 0 | T3 | N0 | M0 | 2A | II | Descending colon | 3.5×3×2 |
| D15A1628-B30-C1 | 2 | 25 | 1 | 76 | 0 | T3 | N1a | M0 | 3B | II | Ascending colon | 8×8×4 |
| D15A1615-B30-C1 | 1 | 61 | 0 | 50 | 1 | T1 | N0 | M0 | 1 | I | Ascending colon | 4×3×3 |
| D15A1616-B30-C1 | 1 | 61 | 0 | 74 | 0 | T3 | N0 | M0 | 2A | I | Ascending colon | 5×2.5×1 |
| D15A1617-B30-C1 | 1 | 61 | 0 | 80 | 1 | T3 | N0 | M0 | 2A | II | Right colon | 8×7×1 |
| D15A1619-B30-C1 | 2 | 61 | 0 | 65 | 0 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 4×3.5×1 |
| D15A1620-B30-C1 | 1 | 61 | 0 | 59 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 4.5×3.5×1.2 |
| D15A1622-B30-C1 | 2 | 61 | 0 | 79 | 1 | T3 | N0 | M0 | 2A | I–II | Descending colon | 4×4×1 |
| D15A1629-B30-C1 | 2 | 61 | 0 | 56 | 1 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 4×3×1 |
| D15A1624-B30-C1 | 2 | 13 | 1 | 76 | 0 | T4a | N1b | M0 | 3B | II | Sigmoid colon | 3.5×3.5×1 |
| D15A1625-B30-C1 | 2 | 60 | 0 | 76 | 0 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 8×6×1 |
| D15A1626-B30-C1 | 1 | 39 | 1 | 63 | 1 | T4a | N1b | M0 | 3B | I–II | Sigmoid colon | 5×3×1.5 |
| D15A1630-B30-C1 | 1 | 60 | 0 | 44 | 0 | T2 | N0 | M0 | 1 | II | Ascending colon | 8×8×4 |
| D15A1663-B30-C1 | 1 | 13 | 1 | 73 | 1 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 5.5×3.5×2 |
| D15A1668-B30-C1 | 1 | 60 | 0 | 66 | 0 | T1 | N1a | M0 | 3A | II | Transverse colon | 7.5×6.5×0.5 |
| D15A1669-B30-C1 | 2 | 1 | 1 | 48 | 0 | T3 | N0 | M0 | 2A | II | Right colon | 8×7×2 |
| D15A1732-B30-C1 | 1 | 59 | 0 | 79 | 1 | T3 | N0 | M0 | 2A | I–II | Ascending colon | 6×3×1 |
| D15A1733-B30-C1 | 1 | 59 | 0 | 55 | 1 | T4a | N1b | M0 | 3B | II–III | Hepatic flexure | 5×4×1 |
| D15A1735-B30-C1 | 2 | 59 | 0 | 65 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 4×3×1 |
| D15A1740-B30-C1 | 2 | 7 | 1 | 73 | 0 | T3 | N1a | M0 | 3B | II | Right colon | 8×5×2.5 |
| D15A1741-B30-C1 | 1 | 59 | 0 | 81 | 1 | T3 | N1a | M0 | 3B | II | Left colon | 8×7×1.5 |
| D15A1742-B30-C1 | 2 | 59 | 0 | 61 | 1 | T2 | N0 | M0 | 1 | I–II | Descending colon | 4.5×3.5×1.5 |
| D15A1745-B30-C1 | 2 | 59 | 0 | 80 | 1 | T3 | N0 | M0 | 2A | II | Ascending colon | 4×3×2 |
| D15A1743-B30-C1 | 2 | 16 | 1 | 65 | 1 | T4b | N1b | M0 | 3C | III | Colon | 6×5×1.3 |
| D15A1744-B30-C1 | 2 | 16 | 1 | 61 | 1 | T4a | N2a | M1a | 4A | II–III | Sigmoid colon | 4×4×3 |
| D15A1756-B30-C1 | 2 | 58 | 0 | 71 | 0 | T3 | N1a | M0 | 3B | II | Sigmoid colon | 3×1.5×1 |
| D15A1758-B30-C1 | 1 | 58 | 0 | 55 | 0 | T3 | N0 | M0 | 2A | II–III | Ascending colon | 11×6×3 |
| D15A1765-B30-C1 | 2 | 21 | 1 | 55 | 1 | T4a | N0 | M0 | 2B | II | Sigmoid colon | 4×2.5×1 |
| D15A1767-B30-C1 | 1 | 58 | 0 | 83 | 1 | T3 | N0 | M0 | 2A | I | Ascending colon | 5×3×2 |
| D15A1762-B30-C1 | 1 | 58 | 0 | 69 | 0 | T4a | N0 | M0 | 2B | II | Transverse colon | 8×5×4 |
| D15A1764-B30-C1 | 58 | 0 | 80 | 0 | T4a | N1a | M0 | 3B | II–III | Ascending colon | 6×5.5×1 | |
| D15A1990-B30-C1 | 1 | 57 | 0 | 43 | 1 | T3 | N0 | M0 | 2A | II | Right colon | 4×3×1.5 |
| D15A1811-B30-C1 | 2 | 57 | 0 | 73 | 1 | T4a | N0 | M0 | 2B | I | Ascending colon | 7×4×1 |
| D15A1813-B30-C1 | 1 | 57 | 0 | 82 | 0 | T3 | N1a | M0 | 3B | II–III | Transverse colon | 4×4×1 |
| D15A1814-B30-C1 | 2 | 57 | 0 | 69 | 0 | T2 | N0 | M0 | 1 | I–II | Ascending colon | 2×2×1.5 |
| D15A1991-B30-C1 | 2 | 57 | 0 | 83 | 1 | T4a | N0 | M0 | 2B | II | Descending colon | 4×2×1 |
| D15A1815-B30-C1 | 2 | 57 | 0 | 46 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 6×6×0.7 |
| D15A1819-B30-C1 | 2 | 57 | 0 | 56 | 0 | T3 | N0 | M0 | 2A | II | Descending colon | 4.5×3.5×1.5 |
| D15A1992-B30-C1 | 56 | 0 | 66 | 1 | T3 | N0 | M0 | 2A | II | Ascending colon | 4×2.5×0.6 | |
| D15A1993-B30-C1 | 2 | 0.4 | 1 | 82 | 1 | T3 | N2b | M0 | 3C | III | Splenic flexure | 7×6×1 |
| D15A1820-B30-P1 | 1 | 56 | 0 | 78 | 0 | T3 | N0 | M0 | 2A | II | Sigmoid colon | 4.5×4×1.5 |
| D15A1836-B30-C1 | 1 | 56 | 0 | 81 | 0 | T4a | N0 | M0 | 2B | II | Colon | 6×5×3.5 |
| D15A1839-B30-C1 | 2 | 56 | 0 | 73 | 1 | T4a | N0 | M0 | 2B | II | Sigmoid colon | 5×5×1.5 |
| D15A1841-B30-C1 | 1 | 56 | 0 | 50 | 0 | T3 | N0 | M0 | 2A | II | Right colon | 6×4×1 |
| D15A1904-B30-C1 | 19 | 1 | 27 | 0 | T4a | N2a | M0 | 3C | III | Descending colon | 4×4×1.5 | |
| D15A1907-B30-C1 | 2 | 35 | 1 | 54 | 1 | T3 | N0 | M0 | 2A | II | Right colon | 5×5×2 |
| D15A1914-B30-C1 | 2 | 55 | 0 | 77 | 0 | T4a | N1a | M0 | 3B | I–II | Splenic flexure | |
| D15A1915-B30-C1 | 1 | 55 | 0 | 55 | 0 | T4a | N1b | M0 | 3B | II | Sigmoid colon | 9×6×2 |
| D15A1917-B30-C1 | 1 | 55 | 0 | 66 | 1 | T2 | N0 | M0 | 1 | II | Sigmoid colon | 2.7×2.2×1.3 |
| D15A1918-B30-C1 | 1 | 42 | 1 | 60 | 1 | T3 | N2b | M0 | 3C | II | Sigmoid colon | 3.5×2×1.5 |
| D15A1919-B30-C1 | 1 | 19 | 1 | 65 | 1 | T3 | N2a | M0 | 3B | II | Sigmoid colon | 5×5×1.8 |
| D15A1921-B30-C1 | 15 | 1 | 56 | 1 | T3 | N1b | M0 | 3B | II | Sigmoid colon | 6×6×2.5 | |
| D15A1923-B30-C1 | 2 | 55 | 0 | 54 | 1 | T4a | N1b | M0 | 3B | II | Sigmoid colon | 6.5×5×2.5 |
| D15A1928-B30-C1 | 1 | 55 | 0 | 52 | 0 | T2 | N0 | M0 | 1 | II | Transverse colon | 5.5×4.5×1.5 |
| D15A1929-B30-C1 | 1 | 23 | 1 | 62 | 0 | T4a | N1b | M0 | 3B | I–II | Ascending colon | 5×4×3 |
| D15A1927-B30-C1 | 2 | 19 | 1 | 67 | 1 | T3 | N1a | M0 | 3B | II | Sigmoid colon | 6×5×1 |
*, CypB (score 0–2 =1, score 3–4 =2); #, OS (event =1); &, Sex (male =1, female =2).
Table S2
| Sample | CypB mRNA expression | OS time (days) | OS status (event =1) | M stage | N stage | T stage | TNM stage | Gender | Age (year) | Neoplasm_subdivision |
|---|---|---|---|---|---|---|---|---|---|---|
| TCGA-3L-AA1B-01 | 13.3798 | 475 | 0 | M0 | N0 | T2 | I | Female | 61 | Cecum |
| TCGA-4N-A93T-01 | 12.6538 | 146 | 0 | M0 | N1b | T4a | IIIB | Male | 67 | Ascending colon |
| TCGA-4T-AA8H-01 | 12.83 | 385 | 0 | MX | N0 | T3 | IIA | Female | 42 | Descending colon |
| TCGA-5M-AAT4-01 | 12.5424 | 49 | 1 | M1b | N0 | T3 | IV | Male | 74 | Ascending colon |
| TCGA-5M-AAT5-01 | 13.5081 | |||||||||
| TCGA-5M-AAT6-01 | 13.9539 | 290 | 1 | M1a | N2b | T4a | IV | Female | 40 | Transverse colon |
| TCGA-5M-AATA-01 | 13.2919 | |||||||||
| TCGA-5M-AATE-01 | 12.5337 | 1,200 | 0 | M0 | N0 | T3 | IIA | Male | 76 | Ascending colon |
| TCGA-A6-2675-01 | 12.1484 | 1,321 | 0 | MX | N0 | T3 | IIA | Male | 78 | Sigmoid colon |
| TCGA-A6-2682-01 | 13.725 | 424 | 1 | M1 | N1 | T4b | IV | Male | 70 | [Discrepancy] |
| TCGA-A6-2684-01 | 13.1669 | 1,127 | 0 | M0 | N0 | T2 | I | Female | 75 | Cecum |
| TCGA-A6-2685-01 | 12.9514 | 1,133 | 0 | M0 | N0 | T3 | IIA | Female | 48 | Sigmoid colon |
| TCGA-A6-2686-01 | 13.1465 | 1,126 | 1 | M0 | N0 | T3 | IIA | Female | 81 | Cecum |
| TCGA-A6-4105-01 | 13.7982 | 442 | 1 | M0 | N0 | T3 | IIA | Male | 79 | Ascending colon |
| TCGA-A6-5656-01 | 13.305 | 1,001 | 0 | M0 | N0 | T2 | I | Male | 74 | Sigmoid colon |
| TCGA-A6-5657-01 | 12.9149 | 962 | 0 | M0 | N1 | T3 | IIIB | Male | 65 | [Discrepancy] |
| TCGA-A6-5659-01 | 13.0106 | 926 | 0 | M0 | N0 | T2 | I | Male | 82 | Cecum |
| TCGA-A6-5660-01 | 12.8449 | 888 | 0 | M0 | N2b | T3 | IIIC | Male | 73 | Cecum |
| TCGA-A6-5661-01 | 13.3289 | 1,020 | 0 | M0 | N0 | T3 | IIA | Female | 80 | Ascending colon |
| TCGA-A6-5662-01 | 13.254 | 718 | 0 | M1 | N2 | T3 | IVA | Male | 46 | Splenic flexure |
| TCGA-A6-5664-01 | 13.7724 | 672 | 0 | MX | N2a | T4a | IIIC | Male | 80 | Cecum |
| TCGA-A6-5665-01 | 13.7718 | 671 | 0 | M0 | N0 | T3 | IIA | Female | 84 | Ascending colon |
| TCGA-A6-5666-01 | 13.8249 | 995 | 0 | M0 | N0 | T4b | IIC | Male | 78 | Sigmoid colon |
| TCGA-A6-5667-01 | 12.7441 | 887 | 0 | MX | N1a | T3 | IIIB | Female | 40 | Sigmoid colon |
| TCGA-A6-6137-01 | 12.8026 | 824 | 0 | M0 | N1c | T3 | IIIB | Male | 55 | Hepatic flexure |
| TCGA-A6-6138-01 | 12.2254 | 685 | 0 | M0 | N0 | T2 | I | Male | 61 | Cecum |
| TCGA-A6-6140-01 | 13.035 | 734 | 0 | M0 | N0 | T3 | IIA | Male | 62 | Descending colon |
| TCGA-A6-6141-01 | 13.3598 | 130 | 0 | M0 | N0 | T3 | IIA | Male | 31 | Cecum |
| TCGA-A6-6142-01 | 13.4184 | 763 | 0 | M1a | N1a | T3 | IVA | Female | 56 | Sigmoid colon |
| TCGA-A6-6648-01 | 12.4873 | 766 | 0 | M1a | N0 | T3 | IVA | Male | 56 | [Discrepancy] |
| TCGA-A6-6649-01 | 12.9726 | 735 | 0 | M0 | N1b | T3 | IIIB | Male | 66 | Hepatic flexure |
| TCGA-A6-6650-01 | 12.5784 | 627 | 0 | M0 | N0 | T3 | IIA | Female | 69 | Cecum |
| TCGA-A6-6651-01 | 13.1779 | 662 | 0 | MX | N1b | T3 | IIIB | Female | 55 | Transverse colon |
| TCGA-A6-6652-01 | 12.7351 | 751 | 0 | M1 | N0 | T3 | IVA | Male | 59 | Sigmoid colon |
| TCGA-A6-6653-01 | 13.8395 | 742 | 0 | M0 | N0 | T2 | I | Male | 82 | Ascending colon |
| TCGA-A6-6654-01 | 13.3981 | 726 | 0 | M0 | N1 | T3 | IIIB | Female | 65 | Descending colon |
| TCGA-A6-6780-01 | 13.771 | 612 | 0 | MX | N0 | T3 | IIA | Male | 74 | [Discrepancy] |
| TCGA-A6-6781-01 | 14.0498 | 598 | 0 | MX | N1b | T4b | IIIC | Male | 43 | Transverse colon |
| TCGA-A6-6782-01 | 13.0313 | 617 | 0 | MX | N0 | T4a | IIB | Male | 82 | Transverse colon |
| TCGA-A6-A565-01 | 13.0484 | 494 | 1 | MX | N2 | T3 | IIIC | Female | 34 | Transverse colon |
| TCGA-A6-A566-01 | 13.5206 | 758 | 1 | M0 | N1 | T4 | IIIB | Female | 55 | Descending colon |
| TCGA-A6-A567-01 | 12.2014 | 1,881 | 1 | M1 | N1 | T3 | IV | Male | 56 | Sigmoid colon |
| TCGA-A6-A56B-01 | 12.4469 | 1,711 | 1 | M0 | N1 | T3 | IIIB | Male | 57 | Sigmoid colon |
| TCGA-A6-A5ZU-01 | 13.256 | 293 | 0 | M0 | N1 | T3 | IIIB | Male | 59 | Transverse colon |
| TCGA-AA-3489-01 | 12.8667 | 214 | 1 | M0 | N0 | T3 | II | Male | 75 | Sigmoid colon |
| TCGA-AA-3492-01 | 13.3061 | 92 | 1 | M0 | N0 | T3 | II | Female | 90 | Ascending colon |
| TCGA-AA-3495-01 | 13.2578 | 1,127 | 0 | M0 | N0 | T2 | I | Male | 79 | Hepatic flexure |
| TCGA-AA-3496-01 | 13.0737 | 31 | 0 | M0 | N0 | T3 | II | Female | 83 | Ascending colon |
| TCGA-AA-3502-01 | 12.9701 | 1,065 | 0 | M0 | N0 | T2 | I | Male | 73 | Transverse colon |
| TCGA-AA-3506-01 | 13.5302 | 1,765 | 0 | M0 | N0 | T2 | I | Male | 77 | Hepatic flexure |
| TCGA-AA-3509-01 | 13.2673 | 1,915 | 0 | M0 | N0 | T3 | II | Female | 54 | Sigmoid colon |
| TCGA-AA-3511-01 | 12.7691 | 212 | 0 | M0 | N0 | T4 | II | Male | 64 | Sigmoid colon |
| TCGA-AA-3526-01 | 14.0539 | 580 | 0 | M0 | N0 | T2 | I | Male | 57 | Sigmoid colon |
| TCGA-AA-3655-01 | 13.1989 | 1,856 | 0 | M0 | N0 | T3 | II | Male | 68 | Sigmoid colon |
| TCGA-AA-3660-01 | 13.0301 | 2,375 | 0 | M0 | N0 | T3 | II | Female | 51 | Sigmoid colon |
| TCGA-AA-3662-01 | 12.9868 | 184 | 0 | M1 | N2 | T4 | IV | Female | 80 | Sigmoid colon |
| TCGA-AA-3663-01 | 14.2051 | 212 | 0 | M0 | N0 | T3 | II | Male | 42 | Cecum |
| TCGA-AA-3675-01 | 13.3136 | 1,431 | 0 | M0 | N0 | T3 | II | Male | 84 | Hepatic flexure |
| TCGA-AA-3685-01 | 13.8864 | 1,127 | 0 | M0 | N0 | T3 | II | Male | 69 | Sigmoid colon |
| TCGA-AA-3697-01 | 12.3795 | 2,587 | 0 | M0 | N0 | T3 | II | Male | 77 | Sigmoid colon |
| TCGA-AA-3712-01 | 13.2138 | M0 | N2 | T3 | III | Male | 65 | Descending colon | ||
| TCGA-AA-3713-01 | 12.8525 | 579 | 0 | M1 | N0 | T3 | IV | Male | 68 | Ascending colon |
| TCGA-AA-A01P-01 | 13.7176 | 1,158 | 1 | M0 | N1 | T3 | III | Female | 80 | Ascending colon |
| TCGA-AA-A01X-01 | 12.6789 | 791 | 0 | M0 | N1 | T2 | III | Female | 80 | Sigmoid colon |
| TCGA-AA-A01Z-01 | 13.3704 | 1,126 | 0 | M0 | N0 | T3 | II | Male | 68 | Ascending colon |
| TCGA-AA-A02K-01 | 12.5992 | 426 | 1 | M1 | N2 | T4 | IV | Male | 50 | Ascending colon |
| TCGA-AA-A02Y-01 | 13.3555 | 1,216 | 0 | M0 | N0 | T2 | I | Male | 73 | Cecum |
| TCGA-AD-5900-01 | 13.3362 | 370 | 0 | MX | N0 | T2 | I | Male | 67 | Ascending colon |
| TCGA-AD-6548-01 | 13.4931 | 650 | 0 | M0 | N0 | T2 | I | Female | 81 | Splenic flexure |
| TCGA-AD-6888-01 | 13.8273 | 472 | 1 | M0 | N1b | T3 | IIIB | Male | 73 | Hepatic flexure |
| TCGA-AD-6889-01 | 15.2878 | 2,532 | 1 | M0 | N0 | T3 | IIA | Male | 76 | Ascending colon |
| TCGA-AD-6890-01 | 13.9953 | 746 | 0 | MX | N0 | T1 | Male | 65 | Ascending colon | |
| TCGA-AD-6895-01 | 13.6519 | 763 | 0 | M0 | N1a | T3 | IIIB | Male | 84 | Cecum |
| TCGA-AD-6899-01 | 12.7928 | 176 | 1 | MX | N2b | T4a | IIIC | Male | 84 | Cecum |
| TCGA-AD-6901-01 | 13.1607 | 682 | 1 | MX | N0 | T3 | Male | 78 | Cecum | |
| TCGA-AD-6963-01 | 12.9269 | 834 | 0 | MX | N0 | T3 | Male | 58 | Ascending colon | |
| TCGA-AD-6964-01 | 13.844 | 331 | 1 | N2b | T4a | Male | 58 | Cecum | ||
| TCGA-AD-6965-01 | 13.391 | 805 | 0 | M0 | N2b | T4a | IIIC | Male | 62 | Cecum |
| TCGA-AD-A5EJ-01 | 14.104 | MX | N0 | T3 | IIA | Female | 74 | Cecum | ||
| TCGA-AD-A5EK-01 | 12.9109 | 500 | 0 | MX | N0 | T2 | I | Male | 51 | Ascending colon |
| TCGA-AM-5820-01 | 13.1072 | 14 | 0 | M1 | N2 | T4a | IVA | Female | 59 | Sigmoid colon |
| TCGA-AM-5821-01 | 13.9842 | 28 | 0 | M0 | N0 | T3 | IIA | Female | 68 | Sigmoid colon |
| TCGA-AU-3779-01 | 12.9971 | M0 | N0 | T3 | IIA | Female | 80 | Rectosigmoid junction | ||
| TCGA-AU-6004-01 | 12.4943 | 824 | 0 | M0 | N0 | T2 | I | Female | 69 | Cecum |
| TCGA-AY-5543-01 | 12.6835 | 1,004 | 0 | M1 | N1a | T3 | IVA | Female | 65 | Ascending colon |
| TCGA-AY-6196-01 | 12.8902 | N2b | T3 | IIIC | Male | 47 | Cecum | |||
| TCGA-AY-6197-01 | 13.1902 | 652 | 0 | N0 | T3 | IIA | Male | 60 | Cecum | |
| TCGA-AY-6386-01 | 13.7194 | 542 | 0 | M0 | N1a | T3 | IIIB | Female | 66 | Cecum |
| TCGA-AY-A54L-01 | 13.4659 | 525 | 0 | M0 | N0 | T2 | I | Female | 74 | Transverse colon |
| TCGA-AY-A69D-01 | 12.8051 | 543 | 0 | M0 | N0 | T3 | IIA | Female | 55 | Transverse colon |
| TCGA-AY-A71X-01 | 13.3791 | 588 | 0 | M0 | N0 | T2 | I | Female | 54 | Cecum |
| TCGA-AY-A8YK-01 | 12.3245 | 573 | 0 | M1 | N2a | T3 | IVA | Male | 44 | Sigmoid colon |
| TCGA-AZ-4313-01 | 14.2427 | 2,310 | 0 | M0 | N0 | T1 | I | Female | 51 | Descending colon |
| TCGA-AZ-4315-01 | 13.733 | 1,776 | 0 | M0 | N0 | T3 | IIA | Male | 61 | Cecum |
| TCGA-AZ-4323-01 | 13.2983 | 43 | 1 | M1 | N2 | T4 | IV | Male | 37 | Cecum |
| TCGA-AZ-4614-01 | 13.5968 | 172 | 1 | M1 | N1 | T4a | IVA | Female | 71 | |
| TCGA-AZ-4615-01 | 13.3781 | 1,002 | 0 | M0 | N1 | T3 | IIIB | Male | 84 | |
| TCGA-AZ-4616-01 | 14.3653 | 156 | 1 | M1 | N2 | T3 | IV | Female | 82 | Cecum |
| TCGA-AZ-4682-01 | 13.873 | 680 | 1 | M1 | N0 | T3 | IVA | Male | 61 | Sigmoid colon |
| TCGA-AZ-4684-01 | 12.7842 | 1,977 | 0 | M1 | N2 | T3 | IVA | Male | 49 | |
| TCGA-AZ-5403-01 | 13.307 | 1,910 | 1 | MX | N0 | T3 | II | Male | 43 | Sigmoid colon |
| TCGA-AZ-5407-01 | 12.756 | 2,683 | 0 | M0 | N0 | T1 | I | Female | 51 | Cecum |
| TCGA-AZ-6598-01 | 13.516 | 1,503 | 1 | MX | N0 | T3 | II | Female | 77 | [Discrepancy] |
| TCGA-AZ-6599-01 | 13.5732 | 206 | 1 | MX | N0 | T2 | I | Male | 72 | Cecum |
| TCGA-AZ-6600-01 | 13.2994 | M1 | N1 | T4 | IV | Male | 64 | Hepatic flexure | ||
| TCGA-AZ-6601-01 | 13.0715 | 3,042 | 1 | M0 | N0 | T3 | II | Male | 68 | Sigmoid colon |
| TCGA-AZ-6603-01 | 12.8988 | 899 | 1 | MX | N1 | T2 | Female | 77 | Sigmoid colon | |
| TCGA-AZ-6605-01 | 13.3891 | 159 | 1 | M0 | N1 | T4 | IIIB | Male | 77 | Ascending colon |
| TCGA-AZ-6606-01 | 12.7094 | 357 | 1 | M1 | N2 | T4 | IV | Male | 81 | Cecum |
| TCGA-AZ-6607-01 | 12.8187 | 97 | 1 | M1 | N2 | T4 | IV | Male | 69 | Sigmoid colon |
| TCGA-AZ-6608-01 | 13.6897 | 59 | 1 | M0 | N1 | T2 | IIIA | Female | 55 | Sigmoid colon |
| TCGA-CA-5254-01 | 14.2203 | 386 | 0 | M0 | N0 | T3 | IIA | Female | 42 | Transverse colon |
| TCGA-CA-5255-01 | 13.548 | 376 | 0 | M0 | N0 | T3 | IIA | Male | 45 | Ascending colon |
| TCGA-CA-5256-01 | 13.6084 | 379 | 0 | M0 | N0 | T3 | IIA | Female | 54 | Hepatic flexure |
| TCGA-CA-5796-01 | 12.7443 | 377 | 0 | M0 | N0 | T3 | IIA | Female | 52 | Ascending colon |
| TCGA-CA-5797-01 | 13.4717 | 383 | 0 | M0 | N0 | T3 | IIA | Male | 56 | Sigmoid colon |
| TCGA-CA-6715-01 | 13.6661 | 383 | 0 | M0 | N1 | T3 | IIIB | Male | 63 | Sigmoid colon |
| TCGA-CA-6716-01 | 13.0409 | 371 | 0 | M0 | N0 | T3 | IIA | Male | 65 | Ascending colon |
| TCGA-CA-6717-01 | 12.8855 | 388 | 0 | M0 | N0 | T3 | IIA | Male | 57 | Ascending colon |
| TCGA-CA-6718-01 | 13.3607 | 306 | 1 | M0 | N0 | T3 | IIA | Male | 46 | Ascending colon |
| TCGA-CA-6719-01 | 12.9851 | 435 | 0 | M0 | N0 | T3 | IIA | Male | 77 | Descending colon |
| TCGA-CK-4947-01 | 13.2784 | 534 | 0 | M0 | N1 | T4 | IIIB | Female | 46 | Sigmoid colon |
| TCGA-CK-4948-01 | 13.296 | 4,502 | 0 | M0 | N1 | T3 | III | Female | 45 | Sigmoid colon |
| TCGA-CK-4950-01 | 13.4453 | 2,599 | 0 | M0 | N1 | T3 | IIIB | Female | 68 | Cecum |
| TCGA-CK-4951-01 | 13.3511 | 2,134 | 1 | M0 | N0 | T3 | IIA | Female | 79 | Ascending colon |
| TCGA-CK-4952-01 | 13.4197 | 475 | 0 | M0 | N2 | T4 | IIIC | Female | 48 | Ascending colon |
| TCGA-CK-5912-01 | 13.2997 | 1,493 | 1 | MX | N0 | T2 | I | Male | 81 | Cecum |
| TCGA-CK-5913-01 | 13.4065 | 1,561 | 0 | MX | N0 | T3 | IIA | Female | 58 | Cecum |
| TCGA-CK-5914-01 | 13.1163 | 304 | 0 | MX | N1 | T3 | IIIB | Male | 81 | Sigmoid colon |
| TCGA-CK-5915-01 | 12.3319 | MX | N0 | T2 | I | Male | 63 | Sigmoid colon | ||
| TCGA-CK-5916-01 | 13.7034 | 643 | 1 | M0 | N0 | T1 | I | Female | 71 | Cecum |
| TCGA-CK-6746-01 | 14.0824 | MX | N0 | T4b | IIB | Female | 84 | Cecum | ||
| TCGA-CK-6747-01 | 13.402 | 2,523 | 0 | MX | N0 | T3 | IIA | Female | 87 | Cecum |
| TCGA-CK-6748-01 | 13.2504 | 58 | 0 | M1 | N1 | T3 | IV | Female | 45 | Sigmoid colon |
| TCGA-CK-6751-01 | 13.8717 | 3,780 | 0 | MX | N0 | T2 | I | Female | 88 | Ascending colon |
| TCGA-CM-4743-01 | 14.6461 | 701 | 0 | M0 | N0 | T3 | IIA | Male | 69 | Hepatic flexure |
| TCGA-CM-4744-01 | 14.4513 | 609 | 0 | M0 | N0 | T2 | I | Male | 69 | Cecum |
| TCGA-CM-4747-01 | 12.6867 | 761 | 0 | M1a | N1b | T4a | IVA | Male | 47 | Cecum |
| TCGA-CM-4751-01 | 12.6197 | 822 | 0 | M0 | N1b | T3 | IIIB | Male | 62 | Cecum |
| TCGA-CM-5344-01 | 13.9646 | 670 | 0 | M0 | N1b | T3 | IIIB | Female | 39 | Sigmoid colon |
| TCGA-CM-5348-01 | 12.6828 | 699 | 0 | M0 | N1a | T3 | IIIB | Male | 72 | Cecum |
| TCGA-CM-5349-01 | 13.4487 | 915 | 0 | M0 | N0 | T3 | IIA | Female | 68 | Cecum |
| TCGA-CM-5860-01 | 13.2636 | 974 | 0 | M0 | N0 | T3 | IIA | Male | 44 | Ascending colon |
| TCGA-CM-5861-01 | 13.9718 | 457 | 0 | M0 | N0 | T3 | IIA | Female | 63 | Cecum |
| TCGA-CM-5862-01 | 13.4638 | 153 | 1 | M1a | N1a | T3 | IVA | Male | 80 | Ascending colon |
| TCGA-CM-5863-01 | 13.1017 | 457 | 0 | M0 | N1b | T3 | IIIB | Female | 60 | Ascending colon |
| TCGA-CM-5864-01 | 13.0053 | 457 | 0 | M0 | N0 | T2 | I | Male | 60 | Cecum |
| TCGA-CM-5868-01 | 13.1335 | 518 | 0 | M1a | N1a | T4a | IVA | Female | 59 | Sigmoid colon |
| TCGA-CM-6161-01 | 13.1885 | 457 | 0 | M0 | N0 | T2 | I | Female | 36 | Sigmoid colon |
| TCGA-CM-6162-01 | 13.1823 | 365 | 0 | M0 | N1a | T3 | IIIB | Female | 48 | Ascending colon |
| TCGA-CM-6163-01 | 12.274 | 427 | 0 | M0 | N0 | T1 | I | Male | 74 | Sigmoid colon |
| TCGA-CM-6164-01 | 13.0871 | 883 | 0 | M0 | N0 | T3 | IIA | Female | 46 | Sigmoid colon |
| TCGA-CM-6165-01 | 12.0513 | 488 | 0 | M0 | N0 | T3 | IIA | Male | 74 | Sigmoid colon |
| TCGA-CM-6166-01 | 13.4988 | 669 | 0 | M0 | N0 | T2 | I | Female | 48 | Ascending colon |
| TCGA-CM-6167-01 | 12.9749 | 456 | 0 | M0 | N2b | T3 | IIIC | Female | 57 | Cecum |
| TCGA-CM-6168-01 | 13.2209 | 395 | 0 | M0 | N0 | T3 | IIA | Female | 84 | Ascending colon |
| TCGA-CM-6169-01 | 12.6992 | 396 | 0 | M0 | N0 | T3 | IIA | Male | 67 | Cecum |
| TCGA-CM-6170-01 | 12.3852 | 457 | 0 | M0 | N0 | T2 | I | Female | 73 | Descending colon |
| TCGA-CM-6171-01 | 14.4106 | 427 | 0 | M0 | N0 | T2 | I | Female | 77 | Ascending colon |
| TCGA-CM-6172-01 | 12.7955 | 335 | 0 | M0 | N1a | T3 | IIIB | Female | 70 | Sigmoid colon |
| TCGA-CM-6674-01 | 13.8921 | 394 | 0 | M0 | N0 | T3 | IIA | Male | 39 | Hepatic flexure |
| TCGA-CM-6675-01 | 13.0063 | 397 | 0 | M1b | N2b | T3 | IVB | Male | 35 | Cecum |
| TCGA-CM-6676-01 | 12.8475 | 337 | 0 | M0 | N0 | T2 | I | Male | 82 | Sigmoid colon |
| TCGA-CM-6677-01 | 12.6611 | 337 | 0 | M0 | N0 | T3 | IIA | Female | 75 | Hepatic flexure |
| TCGA-CM-6678-01 | 13.5735 | 335 | 0 | M1a | N1c | T4a | IVA | Female | 63 | Sigmoid colon |
| TCGA-CM-6679-01 | 13.2632 | 306 | 0 | M0 | N0 | T3 | IIA | Male | 58 | Sigmoid colon |
| TCGA-CM-6680-01 | 12.8609 | 366 | 0 | M0 | N2a | T3 | IIIB | Female | 78 | Cecum |
| TCGA-D5-5537-01 | 13.707 | 1,381 | 1 | MX | N2 | T3 | IIA | Male | 83 | Ascending colon |
| TCGA-D5-5538-01 | 13.5236 | 1,661 | 1 | M0 | N1b | T3 | IIIB | Female | 60 | Cecum |
| TCGA-D5-5539-01 | 13.0102 | 596 | 0 | M0 | N1 | T3 | IIIA | Male | 60 | Ascending colon |
| TCGA-D5-5540-01 | 13.899 | 1,706 | 0 | M0 | N0 | T3 | IIA | Male | 73 | Cecum |
| TCGA-D5-5541-01 | 13.097 | 1,701 | 0 | M0 | N1a | T3 | IIIB | Male | 63 | Sigmoid colon |
| TCGA-D5-6529-01 | 12.8576 | 614 | 0 | M0 | N0 | T3 | IIA | Male | 69 | [Discrepancy] |
| TCGA-D5-6530-01 | 13.5343 | 621 | 0 | M0 | N0 | T2 | I | Male | 53 | [Discrepancy] |
| TCGA-D5-6531-01 | 13.5619 | 540 | 0 | M0 | N0 | T3 | IIA | Male | 75 | Hepatic flexure |
| TCGA-D5-6532-01 | 13.2976 | 555 | 0 | M0 | N0 | T3 | IIA | Male | 61 | Sigmoid colon |
| TCGA-D5-6533-01 | 12.4948 | 775 | 0 | M0 | N0 | T4b | [Discrepancy] | Female | 68 | Transverse colon |
| TCGA-D5-6534-01 | 13.1514 | 1,316 | 0 | M0 | N0 | T3 | IIA | Female | 62 | Ascending colon |
| TCGA-D5-6535-01 | 12.3147 | 460 | 0 | MX | N1 | T3 | IIIB | Female | 80 | Ascending colon |
| TCGA-D5-6536-01 | 13.7486 | 543 | 0 | M0 | N0 | T3 | IIA | Male | 73 | Sigmoid colon |
| TCGA-D5-6537-01 | 13.3376 | 146 | 1 | MX | N1a | T3 | IIIB | Male | 64 | Transverse colon |
| TCGA-D5-6538-01 | 13.2516 | 521 | 0 | M0 | N2 | T3 | IIIB | Female | 79 | Hepatic flexure |
| TCGA-D5-6539-01 | 12.3064 | 380 | 0 | M0 | N0 | T3 | [Discrepancy] | Female | 45 | Transverse colon |
| TCGA-D5-6540-01 | 13.827 | 491 | 0 | M0 | N0 | T2 | I | Male | 66 | Cecum |
| TCGA-D5-6541-01 | 13.2981 | 474 | 0 | M0 | N0 | T3 | IIA | Male | 49 | Splenic flexure |
| TCGA-D5-6898-01 | 12.4218 | 229 | 0 | M0 | N0 | T2 | I | Female | 51 | Sigmoid colon |
| TCGA-D5-6920-01 | 13.3308 | 377 | 0 | M0 | N0 | T3 | IIA | Female | 77 | Sigmoid colon |
| TCGA-D5-6922-01 | 12.3456 | 308 | 0 | M0 | N1 | T3 | IIIA | Male | 76 | Sigmoid colon |
| TCGA-D5-6923-01 | 12.909 | 378 | 0 | M0 | N0 | T2 | I | Male | 57 | Sigmoid colon |
| TCGA-D5-6924-01 | 13.2702 | 435 | 0 | M0 | N0 | T3 | IIA | Male | 68 | Sigmoid colon |
| TCGA-D5-6926-01 | 12.9835 | 275 | 0 | M0 | N1 | T4a | IIIB | Male | 65 | Sigmoid colon |
| TCGA-D5-6927-01 | 13.9284 | M0 | N0 | T3 | IIA | Male | 34 | Transverse colon | ||
| TCGA-D5-6928-01 | 13.1599 | 354 | 0 | M0 | N0 | T3 | IIA | Male | 80 | Ascending colon |
| TCGA-D5-6929-01 | 13.5511 | 408 | 0 | M1 | N1 | T3 | IV | Female | 49 | Sigmoid colon |
| TCGA-D5-6930-01 | 13.9947 | 406 | 0 | M0 | N0 | T3 | IIA | Male | 67 | Ascending colon |
| TCGA-D5-6931-01 | 13.1779 | 365 | 0 | M0 | N2 | T4b | IIIC | Male | 77 | Transverse colon |
| TCGA-D5-6932-01 | 13.1223 | 346 | 0 | M0 | N0 | T3 | IIA | Male | 69 | Transverse colon |
| TCGA-D5-7000-01 | 13.1931 | 312 | 0 | M0 | N0 | T2 | I | Female | 79 | Cecum |
| TCGA-DM-A0X9-01 | 13.3966 | 3,641 | 0 | M0 | N0 | T3 | IIA | Female | 71 | [Discrepancy] |
| TCGA-DM-A0XD-01 | 13.9004 | 743 | 1 | M0 | N0 | T3 | IIA | Male | 65 | [Discrepancy] |
| TCGA-DM-A0XF-01 | 13.5163 | 1,162 | 1 | M0 | N2 | T3 | IIIC | Female | 68 | [Discrepancy] |
| TCGA-DM-A1D0-01 | 12.3348 | 3,974 | 0 | M0 | N0 | T3 | IIA | Female | 79 | Sigmoid colon |
| TCGA-DM-A1D4-01 | 12.6627 | 2,821 | 1 | M0 | N0 | T3 | IIA | Male | 80 | Cecum |
| TCGA-DM-A1D6-01 | 12.2639 | 1,518 | 1 | M0 | N0 | T3 | IIA | Male | 88 | Splenic flexure |
| TCGA-DM-A1D7-01 | 13.3092 | 405 | 1 | M0 | N0 | T3 | IIA | Male | 82 | Sigmoid colon |
| TCGA-DM-A1D8-01 | 12.7949 | 383 | 1 | N1 | T3 | Female | 50 | Ascending colon | ||
| TCGA-DM-A1D9-01 | 12.5488 | 4,270 | 0 | M0 | N0 | T3 | IIA | Female | 67 | Cecum |
| TCGA-DM-A1DA-01 | 13.3777 | 228 | 1 | M0 | N2 | T3 | IIIC | Female | 71 | Cecum |
| TCGA-DM-A1DB-01 | 13.8398 | 1,348 | 1 | M0 | N0 | T3 | IIA | Male | 68 | Sigmoid colon |
| TCGA-DM-A1HA-01 | 12.4432 | 4,000 | 0 | M0 | N2 | T3 | IIIC | Male | 82 | Ascending colon |
| TCGA-DM-A1HB-01 | 14.6197 | 4,126 | 0 | M0 | N1 | T3 | IIIB | Male | 75 | Transverse colon |
| TCGA-DM-A280-01 | 14.2422 | 236 | 1 | M0 | N0 | T3 | IIA | Female | 70 | Ascending colon |
| TCGA-DM-A282-01 | 13.9437 | 4,233 | 0 | M0 | N0 | T3 | IIA | Female | 60 | Hepatic flexure |
| TCGA-DM-A285-01 | 13.4791 | 179 | 1 | M1 | N2 | T3 | IV | Female | 71 | Ascending colon |
| TCGA-DM-A288-01 | 13.5002 | 427 | 1 | M0 | N2 | T3 | IIIC | Male | 68 | Cecum |
| TCGA-DM-A28A-01 | 13.7039 | 805 | 1 | M0 | N2 | T3 | IIIC | Male | 78 | Cecum |
| TCGA-DM-A28C-01 | 12.6539 | 2,475 | 1 | M0 | N0 | T3 | IIA | Male | 74 | Sigmoid colon |
| TCGA-DM-A28E-01 | 13.4291 | 3,648 | 0 | M0 | N0 | T3 | IIA | Female | 72 | Sigmoid colon |
| TCGA-DM-A28F-01 | 13.009 | 1,094 | 1 | M0 | N1 | T3 | IIIB | Male | 73 | Sigmoid colon |
| TCGA-DM-A28G-01 | 13.1773 | 1,849 | 1 | M0 | N0 | T3 | IIA | Male | 75 | Ascending colon |
| TCGA-DM-A28H-01 | 13.1818 | 3,561 | 0 | M0 | N2 | T3 | IIIC | Male | 50 | Cecum |
| TCGA-DM-A28K-01 | 13.9535 | 2,988 | 0 | M0 | N0 | T3 | IIA | Male | 75 | Hepatic flexure |
| TCGA-DM-A28M-01 | 12.7494 | 2,895 | 0 | M0 | N0 | T3 | IIA | Male | 63 | Descending colon |
| TCGA-F4-6459-01 | 12.7603 | 262 | 1 | M0 | N2a | T3 | IIIB | Female | 61 | Sigmoid colon |
| TCGA-F4-6460-01 | 12.8791 | 972 | 1 | M0 | N1 | T3 | IIIB | Female | 51 | Sigmoid colon |
| TCGA-F4-6461-01 | 13.4939 | 338 | 1 | M0 | N2 | T4b | IIIC | Female | 41 | Hepatic flexure |
| TCGA-F4-6463-01 | 13.5646 | 1,087 | 0 | M0 | N0 | T3 | IIA | Male | 51 | Transverse colon |
| TCGA-F4-6569-01 | 13.2891 | 1,087 | 0 | M0 | N0 | T2 | I | Male | 60 | Transverse colon |
| TCGA-F4-6570-01 | 13.4011 | 188 | 1 | M0 | N0 | T3 | IIA | Female | 78 | Transverse colon |
| TCGA-F4-6703-01 | 13.3259 | 1,456 | 0 | M0 | N0 | T3 | IIA | Male | 64 | Ascending colon |
| TCGA-F4-6704-01 | 13.5814 | 47 | 0 | MX | N2b | T3 | IIIC | Male | 60 | Sigmoid colon |
| TCGA-F4-6805-01 | 13.2868 | 1,047 | 0 | M0 | N0 | T3 | IIA | Female | 58 | Descending colon |
| TCGA-F4-6806-01 | 13.355 | 1,260 | 0 | M0 | N0 | T2 | I | Female | 59 | Sigmoid colon |
| TCGA-F4-6807-01 | 12.5661 | 1,309 | 0 | M0 | N2b | T3 | IIIC | Female | 51 | Hepatic flexure |
| TCGA-F4-6808-01 | 13.6601 | 1,024 | 0 | M0 | N0 | T1 | I | Female | 54 | Sigmoid colon |
| TCGA-F4-6809-01 | 12.8493 | 403 | 1 | M1 | N1 | T3 | IVA | Female | 52 | Sigmoid colon |
| TCGA-F4-6854-01 | 12.4674 | 16 | 0 | M0 | N0 | T3 | IIA | Female | 77 | Sigmoid colon |
| TCGA-F4-6855-01 | 13.0115 | 1,442 | 0 | M0 | N0 | T3 | IIA | Female | 70 | Sigmoid colon |
| TCGA-F4-6856-01 | 13.9564 | 1,074 | 0 | M0 | N0 | T2 | I | Male | 45 | Cecum |
| TCGA-F4-6857-01 | 13.0872 | |||||||||
| TCGA-G4-6293-01 | 13.4778 | 4,051 | 0 | M0 | N1 | T3 | III | Female | 49 | Transverse colon |
| TCGA-G4-6294-01 | 13.2546 | 858 | 1 | M1 | N1 | T3 | IV | Male | 75 | Cecum |
| TCGA-G4-6295-01 | 12.6653 | 254 | 0 | M0 | N0 | T3 | II | Female | 70 | Cecum |
| TCGA-G4-6297-01 | 13.6856 | 2,506 | 0 | M1 | N2 | T3 | IV | Female | 55 | Cecum |
| TCGA-G4-6298-01 | 13.3551 | 715 | 1 | MX | N1 | T4a | IIIB | Male | 90 | Cecum |
| TCGA-G4-6299-01 | 13.2354 | 2,268 | 0 | M0 | N2 | T3 | IIIC | Male | 69 | Descending colon |
| TCGA-G4-6302-01 | 13.2234 | 2,047 | 1 | M0 | N0 | T3 | IIA | Female | 90 | Cecum |
| TCGA-G4-6303-01 | 12.5696 | 2,003 | 1 | M1 | N1 | T3 | IV | Female | 54 | Sigmoid colon |
| TCGA-G4-6304-01 | 14.0855 | 1,631 | 0 | M0 | N0 | T4 | IIB | Female | 66 | Transverse colon |
| TCGA-G4-6306-01 | 13.5552 | 1,359 | 0 | M0 | N0 | T2 | [Discrepancy] | Male | 71 | Ascending colon |
| TCGA-G4-6307-01 | 12.7383 | 1,674 | 0 | M0 | N1 | T3 | IIIB | Female | 37 | Sigmoid colon |
| TCGA-G4-6309-01 | 14.0657 | 2,600 | 0 | M0 | N1 | T3 | IIIB | Female | 40 | Sigmoid colon |
| TCGA-G4-6310-01 | 13.0093 | 1,935 | 0 | M0 | N1 | T3 | IIIB | Male | 69 | Cecum |
| TCGA-G4-6311-01 | 12.8804 | 1,199 | 0 | MX | N1 | T3 | III | Male | 80 | Ascending colon |
| TCGA-G4-6314-01 | 13.0451 | 1,093 | 0 | M1 | N2 | T3 | IV | Female | 76 | Cecum |
| TCGA-G4-6315-01 | 13.0198 | 1,883 | 0 | M1 | N1 | T3 | IV | Male | 66 | Descending colon |
| TCGA-G4-6317-01 | 12.7918 | 1,095 | 0 | MX | N2 | T3 | IIIC | Female | 51 | Sigmoid colon |
| TCGA-G4-6320-01 | 13.278 | 804 | 0 | MX | N1 | T3 | III | Male | 73 | Hepatic flexure |
| TCGA-G4-6321-01 | 12.88 | 672 | 0 | MX | N1 | T2 | III | Female | 60 | Cecum |
| TCGA-G4-6322-01 | 14.2467 | 792 | 0 | MX | N1 | T3 | IIIB | Male | 65 | Descending colon |
| TCGA-G4-6323-01 | 13.1754 | 419 | 0 | MX | N0 | Tis | IA | Male | 50 | Cecum |
| TCGA-G4-6586-01 | 13.683 | 1,089 | 0 | M0 | N0 | T3 | IIA | Female | 73 | Ascending colon |
| TCGA-G4-6588-01 | 13.4753 | 796 | 0 | M0 | N0 | T3 | IIA | Female | 58 | Cecum |
| TCGA-G4-6625-01 | 12.8892 | 2,792 | 0 | M0 | N0 | T3 | IIA | Female | 77 | Sigmoid colon |
| TCGA-G4-6626-01 | 12.547 | 1,422 | 1 | M0 | N0 | T3 | IIA | Male | 90 | Ascending colon |
| TCGA-G4-6627-01 | 12.8671 | 2,275 | 0 | M0 | N0 | T3 | IIA | Male | 84 | Ascending colon |
| TCGA-G4-6628-01 | 13.7375 | 2,424 | 0 | M0 | N0 | T2 | I | Male | 78 | Cecum |
| TCGA-NH-A50T-01 | 13.6842 | 553 | 0 | MX | N0 | T3 | IIA | Female | 68 | Splenic flexure |
| TCGA-NH-A50U-01 | 12.8574 | 334 | 1 | M1a | N0 | T4a | IVA | Male | 42 | Cecum |
| TCGA-NH-A50V-01 | 12.4816 | 588 | 0 | M0 | N2a | T3 | IIIB | Male | 69 | Cecum |
| TCGA-NH-A5IV-01 | 13.1119 | 588 | 0 | MX | N0 | T3 | IIA | Female | 90 | Transverse colon |
| TCGA-NH-A6GA-01 | 12.697 | 302 | 1 | MX | N2a | T4a | IIIC | Male | 58 | Ascending colon |
| TCGA-NH-A6GB-01 | 13.4996 | 476 | 0 | MX | N2b | T3 | IIIC | Female | 71 | Transverse colon |
| TCGA-NH-A6GC-01 | 12.5818 | 389 | 0 | M1b | N1b | T4b | IVB | Female | 66 | Descending colon |
| TCGA-NH-A8F7-01 | 12.7693 | 543 | 0 | MX | N0 | T3 | IIA | Female | 53 | Sigmoid colon |
| TCGA-NH-A8F8-01 | 13.432 | 511 | 1 | M1 | N2b | T4a | IV | Male | 79 | Ascending colon |
| TCGA-QG-A5YV-01 | 13.4551 | 1,301 | 0 | MX | N1a | T4b | IIIC | Female | 64 | Sigmoid colon |
| TCGA-QG-A5YW-01 | 13.0881 | 896 | 0 | MX | N2b | T3 | IIIC | Female | 55 | Cecum |
| TCGA-QG-A5YX-01 | 13.1771 | 1,003 | 0 | MX | N0 | T3 | IIA | Female | 61 | Sigmoid colon |
| TCGA-QG-A5Z1-01 | 12.6054 | 256 | 1 | MX | N1b | T3 | IIIB | Male | 71 | Sigmoid colon |
| TCGA-QG-A5Z2-01 | 13.0875 | 952 | 0 | M0 | N0 | T2 | I | Male | 61 | Cecum |
| TCGA-QL-A97D-01 | 12.3149 | 666 | 0 | MX | N0 | T2 | I | Female | 84 | Cecum |
| TCGA-RU-A8FL-01 | 12.7671 | 1,177 | 0 | MX | N2a | T3 | IIIB | Male | 51 | Cecum |
| TCGA-SS-A7HO-01 | 13.7814 | 1,829 | 0 | M0 | N0 | T4a | IIB | Female | 44 | Cecum |
| TCGA-T9-A92H-01 | 12.5539 | 362 | 0 | M0 | N0 | T3 | IIA | Male | 82 | Sigmoid colon |
| TCGA-WS-AB45-01 | 13.4247 | 2,130 | 0 | MX | N0 | T3 | IIA | Female | 52 | Cecum |
Table S3
| #term ID | Term description | False discovery rate | Matching proteins in your network |
|---|---|---|---|
| hsa04514 | Cell adhesion molecules (CAMs) | 0.00072 | CDH1, CDH3, CLDN1, CLDN2, CLDN4 |
| hsa04530 | Tight junction | 0.00085 | CLDN1, CLDN2, CLDN4, MYH11, MYL9 |
| hsa04270 | Vascular smooth muscle contraction | 0.0019 | ACTG2, MYH11, MYL9, PPP1R14A |
| hsa04670 | Leukocyte transendothelial migration | 0.0019 | CLDN1, CLDN2, CLDN4, MYL9 |
| hsa00480 | Glutathione metabolism | 0.002 | GPX2, GPX3, RRM2 |
| hsa05130 | Pathogenic Escherichia coli infection | 0.002 | CDH1, CLDN1, KRT18 |
| hsa04657 | IL-17 signaling pathway | 0.0079 | CCL20, LCN2, MMP1 |
| hsa05160 | Hepatitis C | 0.0185 | CLDN1, CLDN2, CLDN4 |
| hsa05219 | Bladder cancer | 0.0209 | CDH1, MMP1 |
| hsa00590 | Arachidonic acid metabolism | 0.0396 | GPX2, GPX3 |
| HSA-446728 | Cell junction organization | 8.90E-06 | CDH1, CDH3, CLDN1, CLDN2, CLDN4, LIMS2 |
| HSA-421270 | Cell-cell junction organization | 2.73E-05 | CDH1, CDH3, CLDN1, CLDN2, CLDN4 |
| HSA-445355 | Smooth muscle contraction | 3.79E-05 | ACTG2, LMOD1, MYH11, MYL9 |
| HSA-420029 | Tight junction interactions | 0.0014 | CLDN1, CLDN2, CLDN4 |
| HSA-397014 | Muscle contraction | 0.0018 | ACTG2, DES, LMOD1, MYH11, MYL9 |
| HSA-5625740 | RHO GTPases activate PKNs | 0.0079 | MYH11, MYL9, PPP1R14A |
| HSA-416572 | Sema4D induced cell migration and growth-cone collapse | 0.0162 | MYH11, MYL9 |
| HSA-5625900 | RHO GTPases activate CIT | 0.0162 | MYH11, MYL9 |
| HSA-5627117 | RHO GTPases activate ROCKs | 0.0162 | MYH11, MYL9 |
| HSA-5627123 | RHO GTPases activate PAKs | 0.0162 | MYH11, MYL9 |
| HSA-2022854 | Keratan sulfate biosynthesis | 0.0191 | B3GNT3, PRELP |
| HSA-3928663 | EPHA-mediated growth cone collapse | 0.0191 | MYH11, MYL9 |
| HSA-1592389 | Activation of matrix metalloproteinases | 0.0227 | MMP1, MMP7 |
| HSA-195258 | RHO GTPase effectors | 0.0227 | CDH1, MYH11, MYL9, PPP1R14A |
| HSA-3299685 | Detoxification of reactive oxygen species | 0.0227 | GPX2, GPX3 |
| HSA-418990 | Adherens junctions interactions | 0.0227 | CDH1, CDH3 |
| HSA-202733 | Cell surface interactions at the vascular wall | 0.0231 | CEACAM6, EPCAM, MMP1 |
| HSA-1474228 | Degradation of the extracellular matrix | 0.0239 | CDH1, MMP1, MMP7 |
| HSA-1474244 | Extracellular matrix organization | 0.026 | CDH1, CEACAM6, MMP1, MMP7 |
| HSA-2142753 | Arachidonic acid metabolism | 0.0465 | DPEP1, GPX2 |
Acknowledgments
Funding: This research was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2960). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted under the approval of the Institutional Ethics Committee, Beijing Chao-Yang Hospital of Capital Medical University (No. 2018-Research-61). Written informed consent was obtained from the patient for publication of this study and any accompanying images. The study outcomes will not affect the future management of the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Hoffmann H, Schiene-Fischer C. Functional aspects of extracellular cyclophilins. Biol Chem 2014;395:721-35. [Crossref] [PubMed]
- Yao Q, Li M, Yang H, et al. Roles of cyclophilins in cancers and other organ systems. World J Surg 2005;29:276-80. [Crossref] [PubMed]
- Skagia A, Zografou C, Vezyri E, et al. Cyclophilin PpiB is involved in motility and biofilm formation via its functional association with certain proteins. Genes Cells 2016;21:833-51. [Crossref] [PubMed]
- DeBoer J, Madson CJ, Belshan M. Cyclophilin B enhances HIV-1 infection. Virology 2016;489:282-91. [Crossref] [PubMed]
- Lee J, Choi TG, Ha J, et al. Cyclosporine A suppresses immunoglobulin G biosynthesis via inhibition of cyclophilin B in murine hybridomas and B cells. Int Immunopharmacol 2012;12:42-9. [Crossref] [PubMed]
- Terajima M, Taga Y, Chen Y, et al. Cyclophilin-B Modulates Collagen Cross-linking by Differentially Affecting Lysine Hydroxylation in the Helical and Telopeptidyl Domains of Tendon Type I Collagen. J Biol Chem 2016;291:9501-12. [Crossref] [PubMed]
- Li T, Guo H, Zhao X, et al. Gastric Cancer Cell Proliferation and Survival Is Enabled by a Cyclophilin B/STAT3/miR-520d-5p Signaling Feedback Loop. Cancer Res 2017;77:1227-40. [Crossref] [PubMed]
- Ray P, Rialon-Guevara KL, Veras E, et al. Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J Clin Invest 2012;122:1734-41. [Crossref] [PubMed]
- Kim Y, Jang M, Lim S, et al. Role of cyclophilin B in tumorigenesis and cisplatin resistance in hepatocellular carcinoma in humans. Hepatology 2011;54:1661-78. [Crossref] [PubMed]
- Fang F, Flegler AJ, Du P, et al. Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer. Am J Pathol 2009;174:297-308. [Crossref] [PubMed]
- Ray P, Sullenger BA, White RR. Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: cyclophilin B and its posttranslational modifications. Nucleic Acid Ther 2013;23:435-42. [Crossref] [PubMed]
- Kim K, Kim H, Jeong K, et al. Release of overexpressed CypB activates ERK signaling through CD147 binding for hepatoma cell resistance to oxidative stress. Apoptosis 2012;17:784-96. [Crossref] [PubMed]
- Xiong L, Ding L, Ning H, et al. CD147 knockdown improves the antitumor efficacy of trastuzumab in HER2-positive breast cancer cells. Oncotarget 2016;7:57737-51. [Crossref] [PubMed]
- Li X, Lv L, Zheng J, et al. The significance of LRPPRC overexpression in gastric cancer. Med Oncol 2014;31:818. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Acevedo VD, Gangula RD, Freeman KW, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell 2007;12:559-71. [Crossref] [PubMed]
- Guo L, Han Y. Surgery combined with local 5-aminolevulinic acid-photodynamic therapy on skin cancer and its effect on the expression of cyclophilin A, cyclophilin B and CD147. Oncol Lett 2017;14:1449-54. [Crossref] [PubMed]
- Choi JW, Schroeder MA, Sarkaria JN, et al. Cyclophilin B supports Myc and mutant p53-dependent survival of glioblastoma multiforme cells. Cancer Res 2014;74:484-96. [Crossref] [PubMed]
- Meng DQ, Li PL, Xie M. Expression and role of cyclophilin B in stomach cancer. Genet Mol Res 2015;14:5346-54. [Crossref] [PubMed]
- Jeong K, Kim K, Kim H, et al. Hypoxia induces cyclophilin B through the activation of transcription factor 6 in gastric adenocarcinoma cells. Oncol Lett 2015;9:2854-8. [Crossref] [PubMed]
- Jeong K, Kim H, Kim K, et al. Cyclophilin B is involved in p300-mediated degradation of CHOP in tumor cell adaptation to hypoxia. Cell Death Differ 2014;21:438-50. [Crossref] [PubMed]
- Bingham V, McIlreavey L, Greene C, et al. RNAscope in situ hybridization confirms mRNA integrity in formalin-fixed, paraffin-embedded cancer tissue samples. Oncotarget 2017;8:93392-403. [Crossref] [PubMed]
- Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22-9. [Crossref] [PubMed]
- Choi TG, Nguyen MN, Kim J, et al. Cyclophilin B induces chemoresistance by degrading wild-type p53 via interaction with MDM2 in colorectal cancer. J Pathol 2018;246:115-26. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Fang F, Zheng J, Galbaugh TL, et al. Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells. J Mol Endocrinol 2010;44:319-29. [Crossref] [PubMed]
- Oh Y, Jeong K, Kim K, et al. Cyclophilin B protects SH-SY5Y human neuroblastoma cells against MPP(+)-induced neurotoxicity via JNK pathway. Biochem Biophys Res Commun 2016;478:1396-402. [Crossref] [PubMed]
- Arabzadeh A, Quail DF. Myosin II in Cancer Cells Shapes the Immune Microenvironment. Trends Mol Med 2019;25:257-9. [Crossref] [PubMed]
- Ouderkirk JL, Krendel M. Non-muscle myosins in tumor progression, cancer cell invasion, and metastasis. Cytoskeleton (Hoboken) 2014;71:447-63. [Crossref] [PubMed]
- Derynck R, Weinberg RA. EMT and Cancer: More Than Meets the Eye. Dev Cell 2019;49:313-6. [Crossref] [PubMed]
- Saitoh M. Involvement of partial EMT in cancer progression. J Biochem 2018;164:257-64. [Crossref] [PubMed]


