
Tumor-derived exosomes facilitate tumor cells escape from drug therapy in clear cell renal cell carcinoma
Introduction
Clear cell renal cell carcinoma (ccRCC) accounts for 70% of total renal carcinomas (1). The death rate remains high after metastasis, especially osseous metastasis, pulmonary metastasis, which occurs roughly 15% of total cases (2). Vascular endothelial growth factor (VEGF), raf kinase, and mammalian target of rapamycin (mTOR) inhibitors are the prevalent clinical drugs that have been utilized to treat ccRCC (3,4). More than 50% of ccRCC patients have impaired mTOR signaling pathway (2). Interferon (IFN) are often combined with other drugs to improve the therapeutic efficacy of these inhibitors (1). However, the mechanisms of tumor drug resistance in patients, particularly metastasis and relapse, remains unknown. A large number of hypotheses have been proposed, such as immune escape (5), tumors cell mutant (6,7), vesicle trafficking (8), and microenvironment unbalance (9). Recently, a number of studies indicate that extracellular vesicles, exosomes, play a crucial role in cell communication (10,11).
Exosomes are small extracellular bilayered vesicles, with a diameter ranging between 30 to 150 nm. The biogenesis and release processes of exosome involve a series of proteins, signaling pathways, and organelles (12).
Serum extracellular vesicles (EVs) concentration was found to be higher in hepatocellular carcinoma than in all the other groups, while no evidence to show whether there was the alleviation of EVs in ccRCC. A large number of studies show tumor-derived exosomes (TEXs) could allow tumor cells to escape immune supervision and ultimately immune system mediated apoptosis (12). TEXs can also impact innate immune response via impairing IFN signaling pathway (13). For example, programmed cell death protein 1 is ineffective, due to the adverse effects of exosomes from tumor cells (14). Previous results indicate ccRCC has a higher activation of the mTOR signaling pathway (15), ERK signaling pathway (16,17), or STAT signaling pathway (18). Nevertheless, the evidence showing that TEXs could facilitate ccRCC drug resistance via enhancement of these signal pathways is still limited. Here, we propose that TEXs facilitate ccRCC to resist drug therapy via mTOR-ERK-STAT-NF-κB signaling pathway.
Methods
Samples collect
All tissue samples were collected from ccRCC patients. The control para-carcinoma tissues were gotten from cancer site that more than 2 centimeters away from the same patients.
Reagents
All antibodies were purchased from Cell Signaling Technology (Danvers, USA), Abcam (Cambridge, USA) and Santa Cruz Biotechnology (Santa Cruz, USA). VDAC1 (20B12AF2) (Cat. ab14734) was purchased from Abcam. STAT3 (F-2) (Cat. sc-8019), p-STAT3 antibody (Tyr 705) (Cat. sc-7993), ALIX (3A9) (Cat. Sc-53538), and CD63 (H193) (Cat. Sc-15363) were purchased from Santa Cruz Biotechnology. Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb (Cat. 4370), p44/42 MAPK (ERK1/2) (137F5) Rabbit mAb (Cat. 4695), Phospho-GSK-3β (Ser9) (D85E12) XP® Rabbit mAb (Cat. 5558), GSK-3β (D5C5Z) XP® Rabbit mAb (Cat. 12455), Phospho-NF-κB p65 (Ser536) (93H1) Rabbit mAb (Cat. 3033S), NF-κB p65 (D14E12) XP® Rabbit mAb (Cat. 8242S), phospho-IκBα (Ser32) (14D4) Rabbit mAb (Cat. 2859S), IκBα antibody (Cat. 29242S), phospho-mTOR (Ser2448) antibody (Cat. 2971S), mTOR antibody (Cat. 2972S) were all purchased from CST.
Cell culture
HK-2 cells from normal human kidney tissue-were used as the control for the production of exosomes. 769-P and ACHN cells are epithelial cells, from renal cell adenocarcinoma. 769-P cells were used to produce TEXs. The growth of ACHN cells was inhibited by human IFN, and are suitable for antiproliferative studies. All cell lines were provided by Dr. Qianqian Shi (The Third Affiliated Hospital of Soochow University). HK-2, 769-P, and ACHN cells were cultured in RMPI 1640 medium (Hyclone, GE Lifesciences, USA) with 10% fetal calf serum (FBS) (PAN, Germany) and 100 U/mL Penicillin and 100 µg/mL Streptomycin (Beyotime, China).
Cell proliferation assay
ACHN cells were passaged into 96-well plate with the concentration of 10,000/mL. Cells were treated with 5 µM INFα, 5 µM BP-1-102, 2.5 µM rapamycin, TEXs, INFα + TEXs, BP-1-102 + TEXs, and rapamycin + TEXs for 48 h, respectively. Then detected cell viability follow the CCK8 kit (CK04, Dojindo, Japan) protocol.
RNA extraction
Total RNA was extracted from tissue using a kit (R0027, Beyotime, China). The RNA concentration was measured by NanoDrop Lite (Thermo Fisher Scientific, USA), before reverse transcription.
Real-time PCR
cDNA was generated from RNA, followed the PrimeScriptTM RT reagent Kit (Perfect Real Time) (Cat. RR037A, Takara, Japan) protocol. Real-time PCR primers are listed in Table 1.
Table 1
| Name of primer | Sequence from 5' to 3' |
|---|---|
| F-ALIX | GCCAGAGAACCTAGTGCTCCTT |
| R-ALIX | GGCTTAGTAGGCGGCATGGT |
| F-CD63 | GCGGTGGAAGGAGGAATGA |
| R-CD63 | AGAGACAGAAAGATGGCAAACG |
| F-RAB27A | CCTCAATGTCAGAAACTGGATAAG |
| R-RAB27A | AGTGCTATGGCTTCCTCCTC |
| F-EXOC6B | GCTGTGTCTTCCAGTCCTAGAGAT |
| R-EXOC6B | TAGTGGCTTACTTGAGGCAGGTAG |
| F-STAT3 | GGAGAAGGACATCAGCGGTAAGA |
| R-STAT3 | TAGACCAGTGGAGACACCAGGATA |
| F-STAT5A | GCTTGTGTTCCAGGTGAAGACTC |
| R-STAT5A | GTGGCTGCTGCTGTTGTTGAA |
| F-SIRT2 | TCAAGCCAACCATCTGTCACTACT |
| R-SIRT2 | CCTCCACCAAGTCCTCCTGTTC |
| F-LOXL3 | GGCCCGTGTCCGTCTAAAG |
| R-LOXL3 | TCCAGAGCAGCGAACTTCAC |
| F-ACTIN | TCCCTGGAGAAGAGCTACG |
| R-ACTIN | GTAGTTTCGTGGATGCCACA |
| F-HGS | CTCCTGTTGGAGACAGATTGGG |
| R-HGS | GTGTGGGTTCTTGTCGTTGAC |
| F-TSG101 | ATGGCTACTGGACACATACCC |
| R-TSG101 | GCGGATAGGATGCCGAAATAG |
| F-SNF8 | AGCCCAGATGTCAAAGCAGTT |
| R-SNF8 | CACGGTGTGATCCATATTGAGC |
| F-CHMP4A | GGGACCAAGAATAAGAGAGCTGC |
| R-CHMP4A | TGAAACTCCAGGGTGGATAATGT |
| F-CHMP4B | AGAAGCACGGCACCAAAAAC |
| R-CHMP4B | GCTGGAACTCGATGGTTGATAAT |
| F-CHMP4C | ACTCAGATTGATGGCACACTTTC |
| R-CHMP4C | GCTGCAAAGCCCATGTTCC |
| F-VPS4A | CCACGCTATCAAGTATGAGGC |
| R-VPS4A | CCGTGTTTCTCTTTGCTTCGTA |
| F-VTA1 | CTCCCCGCACAGTTCAAGAG |
| R-VTA1 | AACGACAGTAATAAGCCACCAC |
| F-Syntenin | CTGCTCCTATCCCTCACGATG |
| R-Syntenin | GGCCACATTTGCACGTATTTCT |
| F-Syndecan | ACGGCTATTCCCACGTCTC |
| R-Syndecan | TCTGGCAGGACTACAGCCTC |
| F-USP8 | AAGGAGCAATCACAGCAAAGG |
| R-USP8 | CTGCATTCTTCGAGCATCCATTA |
| F-STAMBP | AGCTGGGTAGTGCGGTAGAG |
| R-STAMBP | TGCCATTCGGATAATCTCAACTC |
| F-ATP6V1H | GGCGCCTCTGTCATTCTACT |
| R-ATP6V1H | CCAAGTAGGCGTCTCCTGTC |
| F-MCOLN1 | GCTGTGACATTCCGGGAAGA |
| R-MCOLN2 | ACCACGGACATACGCATACC |
| F-TFEB | AGAGAATGATGCCTCCGCAC |
| R-TFEB | ATGCGCAACCCTATGCGT |
Exosomes exclusion and exosomes isolation
In order to starve exosomes from fetal bovine serum (FBS), exosomes were depleted FBS via ultracentrifuge at least 16 h at 100,000 g at 4 °C. Exosomes were isolated from cultured supernatant of 769-P and HK-2 cells, the steps as follows: centrifugation at 300 g for 10 min at 4 °C to exclude debris and dead cells, then transferred supernatant to a new tube, centrifuged at 2,000 g for 10 min at 4 °C to exclude some large particles and debris, after that, transferred supernatant to a new tube, centrifuged at 100,000 g for at least 70 min to precipitate exosomes. Discarded supernatants and resuspended sediment with pre-chilled PBS and spun down at 100,000 g for at least 70 min at 4 °C. Finally, these were resuspended with 50 µL PBS to obtain pure exosomes. Exosomes were stored at −20 °C.
Nanoparticle tracking analysis (NTA) of exosomes
NTA is one of the classical and convenient methods to analyze the morphological features of exosomes, which has been widely used in exosomes analysis (19). Isolated exosomes were analyzed via NanoSight NS300 (Malvern, England) after diluted 1,000 to 5,000 times.
Western blot
Cells were seeded a day before treatment. By the 60–80% confluence, the medium was replaced with RMPI containing 2% FBS, followed by treatment with IFNα (Cat. 11200-2, R&D, USA), STAT3 inhibitor 4-(N-(4-Cyclohexylbenzyl)-2-(2,3,4,5,6-pentafluoro-N-methylphenylsulfonamido)acetamido)-2-hydroxybenzoic acid (BP-1-102) (Cat. HY-100493, MCE, USA), mTOR inhibitor rapamycin (Cat. HY-10219, MCE, USA), IFNα and TEXs, BP-1-102 and TEXs, rapamycin and TEXs for 48 h, respectively. Then harvested cells, whole cell lysate (WCL) was analyzed via Western blot.
Statistical analysis
All experiments were repeated triple minimum. Data was expressed mean ± standard error of the mean (SEM) and analyzed via Prism GraphPad 7.0. P value less than 0.05 was rendered statistically significant. All data was analyzed by Student’s t-test or one-way ANOVA.
Results
TEXs secretion was in a high level in renal cancer cells
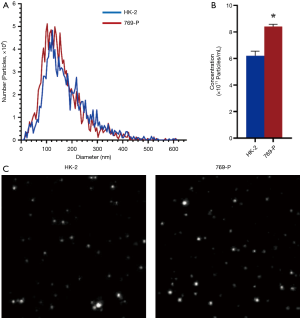
To investigate the exosomal secretion in renal cancer and normal cells, we isolated exosomes from the renal cancer cells 769-P and compared them to the normal renal cell line HK-2. From the results, renal cancer cells’ exosome secretion was higher than normal cells, the mean number of exosomes in HK-2 and 769-P cells were 6.2×1011/mL, 8.2×1011/mL, respectively (Figure 1A,B), meaning there were 32.3% more exosomes secreted by the cancerous cell line. Interesting, no difference in exosome size (Figure 1A,C) was found as the peak analysis size of HK-2-derived exosomes and 769-P-derived exosomes were 118.7 nm and 114.4 nm, respectively.
Exosome biogenesis and transportation mRNA expression upregulated in ccRCC tissue
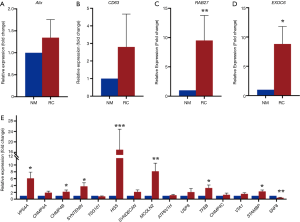
To assess the biogenesis of exosomes, ALIX, CD63, and RAB27A mRNA were analyzed via real-time PCR. Data indicated that all mRNA expression in ccRCC tissues were upregulated (Figure 2A,B,C). EXOC6B, which is involved in vesicle transportation, was also upregulated in cancer tissues (Figure 2D). Next, we wanted to confirm which steps occurred from exosome biogenesis to release. We selected genes of endosomal sorting complexes required for transport (ESCRT), which is the key step of biogenesis of exosomes and lysosome and ubiquitination related genes: VPS4A, CHMP4A/B/C, SYNTENIN, TSG101, HGS, SYNDECAN, MCOLN2, ATP6VH1, USP8, TFEB, VTA1, STAMBP, SNF8. Our results indicated that ESCRT-0, ESCRT-I, ESCRT-III, and VPS4 complex are upregulated except for ESCRT-II (SNF8) while no difference detected in lysosome and ubiquitination genes (Figure 2E).
Gene expression involved in renal cancer development
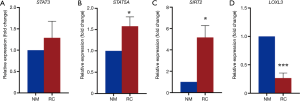
Then we investigated tumor related gene expression. Results showed that STAT3 mRNA was upregulated in tumor tissues (Figure 3A), which is in accordance with previous research indicating the oncogenic role of STAT3 (20). STAT5A, which belongs to the same family of STAT3, was also upregulated in cancer tissues (Figure 3B). Other tumor related genes like SIRT2 also had the similar result as STAT3 (Figure 3C). While LOXL3, which inhibits the activation of STAT3 as we reported (21), was downregulated (Figure 3D). These results suggest that exosomes suppress anti-tumor related gene expression while increase tumor enhance gene expression.
TEXs promoted tumor cells resistant to antitumor drug
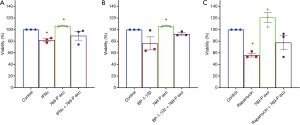
Next, to assess TEXs’ function, ACHN cells treated with mTOR inhibitor rapamycin, STAT3 inhibitor BP-1-102, or IFNα or co-treated with mTOR inhibitor rapamycin and TEXs, STAT3 inhibitor BP-1-102 and TEXs, or IFNα and TEXs, respectively. After ACHN cells were treated with IFNα, BP-1-102, or rapamycin for 48 h, all groups’ viability declined. This effect was elevated if TEXs were added into the supernatant. Studies also showed that TEXs alleviated the death of renal cells (Figure 4A,B,C). These results suggest that TEXs maintain tumor cells survival and resistant to drug treatment.
TEXs enhanced mTOR-ERK1/2-STAT-NF-κB signaling
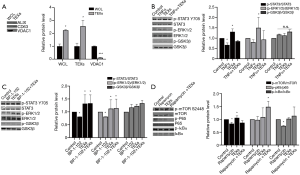
To investigate the mechanisms that TEXs maintained tumor cells’ resistant to antitumor drug, exosomes were confirmed via marker CD63 and ALIX, also confirmed by a mitochondrial marker VDAC1, which was rendered as the negative marker (Figure 5A). Western blot results showed TEXs enhanced STAT3 Y705 phosphorylation (Figure 5B), ERK1/2 phosphorylation (Figure 5C), p65 phosphorylation (Figure 5D), mTOR S2448 phosphorylation (Figure 5D). These results revealed that TEXs facilitated tumor cells escape from drug therapy through mTOR-ERK1/2-STAT-NF-κB signal pathway.
Discussion
Higher concentration of exosomes in tumor patients have been reported previously (22), and our results provide evidence that renal tumor cells release more exosomes compared to normal renal cells in vitro.
ESCRT, which contains ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and VPS4 complex (23), is the crucial first step in the formation of multivesicular bodies (MVBs) which are somewhat precursors to exosomes (24). CD63, ALIX and RAB27A are the common markers of the formation of exosomes (25). We found the alleviation of exosomes, and then we confirmed the upregulation of ESCRT-0, ESCRT-I, ESCRT-III, and VPS4 complex except for ESCRT-II (SNF8). No obvious difference of lysosome and ubiquitination, which involved in degradation of exosomes.
STAT3 was discovered as being important in the regulation of protein utilization in cancer tissues, and as an oncogene (26). Our previous results proved that lysyl oxidase like 3 (Loxl3) negatively regulates STAT3 and affects the proliferation of cells (21). Our results are consistent with previous results that STAT3 is activated in tumor cells (20) as Loxl3 expression is downregulated. The expression of STAT5 and SIRT2, which are related to the proliferation of cells, are upregulated, meaning the proteins involved in enhancing proliferation are upregulated while LOXL3, which suppressed the proliferation, is downregulated.
TEXs act as an important mediator that assist tumor cells survival (27). Our results confirmed that TEXs contribute to the proliferation. But more studies need to be done on detailed components which play the pivotal role in the whole physiological process.
Finally, we found the mechanism of drug resistance in renal cancer occurring via mTOR-ERK-STAT-NF-κB signaling pathways. Previous results proved the importance of mTOR signal pathway, ERK signal pathway, STAT signal pathway, and NF-κB pathway (28-30), but lacked the documents of TEXs. Our results provide an insight of TEXs on ccRCC immune escape.
TEXs have a huge prospect in future diagnosis and therapy because of the detection kit had been using in clinical diagnoses. Glypican-1, which is component of exosomes, could be used as an early marker of pancreatic cancer (31). We still must use conventional methods to isolate exosomes from serum, supernatant, urine, and other fluids so far. A variety of unknown particles, some lipid particles, like low density lipoprotein, will still cause interference with these assays. We have been seeking an advanced approach to detect exosomes with high sensitivity and specificity. In renal carcinoma, miRNA might be good markers for future diagnose (32). If we could confirm ideal markers of exosomes secretion, which can be isolated from urine, and can be used in diagnosis. However, the mechanism of exosomes secretion to outer cells, which would be critical to find a specific target that regulates exosomes biogenesis and transportation and release, is still needs to be uncovered. We did not detect if there are any proteins, DNA, RNA or other substrates in exosomes which play a key role in metabolism process alone or synergistic. So, specific components still need to be elucidated.
Conclusions
From this research, we proposed a hypothesis and documented that TEXs assist tumor cells to escape from immune killing and keep from drug damage via mTOR-ERK-STAT-NF-κB pathway. In the future, exosomes could be used as a potential target and/or vector for tumor therapy.
Acknowledgments
We appreciate Dr. Ying Xie, Dr. Sneha Damal Villiva, Dr. Andrew Widjaja, and Dr. Pete Zushin for their review of the manuscript.
Funding: This study was funded by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2246). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Institutional Ethics Committee of the Third Affiliated Hospital of Soochow University (No. 20170025) and adhered to the principles in the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from each patient before tissue collection for experimentation.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lalani AA, McGregor BA, Albiges L, et al. Systemic Treatment of Metastatic Clear Cell Renal Cell Carcinoma in 2018: Current Paradigms, Use of Immunotherapy, and Future Directions. Eur Urol 2019;75:100-10. [Crossref] [PubMed]
- Shingarev R, Jaimes EA. Renal cell carcinoma: new insights and challenges for a clinician scientist. Am J Physiol Renal Physiol 2017;313:F145-54. [Crossref] [PubMed]
- Pal K, Madamsetty VS, Dutta SK, et al. Synchronous inhibition of mTOR and VEGF/NRP1 axis impedes tumor growth and metastasis in renal cancer. NPJ Precis Oncol 2019;3:31. [Crossref] [PubMed]
- Dorff TB, Goldkorn A, Quinn DI. Targeted therapy in renal cancer. Ther Adv Med Oncol 2009;1:183-205. [Crossref] [PubMed]
- Diegmann J, Junker K, Loncarevic IF, et al. Immune escape for renal cell carcinoma: CD70 mediates apoptosis in lymphocytes. Neoplasia 2006;8:933-8. [Crossref] [PubMed]
- Noonan HR, Metelo AM, Kamei CN, et al. Loss of vhl in the zebrafish pronephros recapitulates early stages of human clear cell renal cell carcinoma. Dis Model Mech 2016;9:873-84. [Crossref] [PubMed]
- Sun C, Zhao C, Li S, et al. EZH2 Expression is increased in BAP1-mutant renal clear cell carcinoma and is related to poor prognosis. J Cancer 2018;9:3787-96. [Crossref] [PubMed]
- Campion CG, Zaoui K, Verissimo T, et al. COMMD5/HCaRG Hooks Endosomes on Cytoskeleton and Coordinates EGFR Trafficking. Cell Rep 2018;24:670-684.e7. [Crossref] [PubMed]
- Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. [Crossref] [PubMed]
- Prieto D, Sotelo N, Seija N, et al. S100-A9 protein in exosomes from chronic lymphocytic leukemia cells promotes NF-kappaB activity during disease progression. Blood 2017;130:777-88. [Crossref] [PubMed]
- Wang Q, Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun 2017;8:709. [Crossref] [PubMed]
- Becker A, Thakur BK, Weiss JM, et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016;30:836-48. [Crossref] [PubMed]
- Gao L, Wang L, Dai T, et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat Immunol 2018;19:233-45. [Crossref] [PubMed]
- Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382-6. [Crossref] [PubMed]
- Zheng Z, Cai Y, Chen H, et al. CXCL13/CXCR5 Axis Predicts Poor Prognosis and Promotes Progression Through PI3K/AKT/mTOR Pathway in Clear Cell Renal Cell Carcinoma. Front Oncol 2019;8:682. [Crossref] [PubMed]
- Chen CM, Hsieh SC, Lin CL, et al. Alpha-Mangostin Suppresses the Metastasis of Human Renal Carcinoma Cells by Targeting MEK/ERK Expression and MMP-9 Transcription Activity. Cell Physiol Biochem 2017;44:1460-70. [Crossref] [PubMed]
- Fang J, Wang H, Cheng G, et al. AB109. Downregulation of tNASP inhibits proliferation through regulating cell cycle-related proteins and inactive ERK/MAPK signal pathway in renal cell carcinoma cells. Transl Androl Urol 2016;5:AB109. [Crossref]
- Brown RE, Buryanek J, Tammisetti VS, et al. Morphoproteomics and biomedical analytics confirm the mTORC2/Akt pathway as a resistance signature and activated ERK and STAT3 as concomitant prosurvival/antiapoptotic pathways in metastatic renal cell carcinoma (RCC) progressing on rapalogs: Pathogenesis and therapeutic options. Oncotarget 2016;7:41612-21. [Crossref] [PubMed]
- Vestad B, Llorente A, Neurauter A, et al. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: a variation study. J Extracell Vesicles 2017;6:1344087. [Crossref] [PubMed]
- Herrmann A, Lahtz C, Song J, et al. Integrin α6 signaling induces STAT3-TET3-mediated hydroxymethylation of genes critical for maintenance of glioma stem cells. Oncogene 2020;39:2156-69. [Crossref] [PubMed]
- Ma L, Huang C, Wang XJ, et al. Lysyl Oxidase 3 Is a Dual-Specificity Enzyme Involved in STAT3 Deacetylation and Deacetylimination Modulation. Mol Cell 2017;65:296-309. [Crossref] [PubMed]
- Zhou Y, Xia L, Lin J, et al. Exosomes in Nasopharyngeal Carcinoma. J Cancer. 2018;9:767-77. [Crossref] [PubMed]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell 2011;21:77-91. [Crossref] [PubMed]
- Dreyer F, Baur A. Biogenesis and Functions of Exosomes and Extracellular Vesicles. Methods Mol Biol 2016;1448:201-16. [Crossref] [PubMed]
- Hassanpour M, Cheraghi O, Akbarzadeh M, et al. CD63-Alix-Rab27a exosome axis is identically influenced in Chediak-Higashi syndrome. Comp Clin Path 2016;25:1313-6. [Crossref]
- Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell 1999;98:295-303. [Crossref] [PubMed]
- Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol 2011;2011:842849.
- Yadav SS, Buryanek J, Brown RE, et al. Morphoproteomics of mTORC2 pathway as resistance signature and activated ERK and STAT3 prosurvival/antiapoptotic pathways in metastatic renal cell carcinoma (mRCC) progressing on rapalogs. J Clin Oncol 2013;31:abstr e15548.
- Chen YC, Chien LH, Huang BM, et al. Aqueous Extracts of Toona sinensis Leaves Inhibit Renal Carcinoma Cell Growth and Migration Through JAK2/stat3, Akt, MEK/ERK, and mTOR/HIF-2 alpha Pathways. Nutr Cancer 2016;68:654-66. [Crossref] [PubMed]
- Zhang M, Pan Y, Jiang R, et al. DKC1 serves as a potential prognostic biomarker for human clear cell renal cell carcinoma and promotes its proliferation, migration and invasion via the NFkappaB pathway. Oncol Rep 2018;40:968-78. [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Wang X, Wang T, Chen C, et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]


