
RGMB-AS1/miR-4428/PBX1 axis drives the progression of cervical cancer
Introduction
CC has the fourth highest morbidity and mortality in cancers (1). This year has seen an increase of about 570,000 diagnoses and 310,000 deaths of CC cases in the world (2). Although CC treatment has been developed for the patients, the prognosis of CC patients is still unoptimistic (3). Hence, mechanism in CC is imperative to be understood so that the therapeutic method for CC could be improved.
Long non-coding RNA (lncRNA) is a newly proposed non-coding RNA (ncRNA) whose length is over 200 nucleotides, and is engaged in regulating cancer progression (4,5). For example, lncRNA HOXD-AS1 enhances the proliferative and invasive capacity of melanoma cells through inhibiting RUNX3 expression (6). Moreover, lncRNA SUMO1P3 facilitates tumor growth, distant metastasis and angiogenesis in colon cancer (7). ZEB1-AS1 shows oncogenic behaviors in gastric cancer and predicts an unfavorable prognosis (8). Numerous recent reports have demonstrated that lncRNA containing miRNA response element (MRE) elicit competitive endogenous RNA (ceRNA) function through sequestering target miRNAs at the MREs (9-11). Interestingly, various lncRNAs functioning as ceRNA are largely documented in CC. For instance, LINC00473 promotes CC tumorigenesis by targeting miR-34a and preventing ILF2 degradation (12). SNHG16, a sponge of miR-216-5p, contributes to the migration and invasion of CC cells through regulating ZEB1 (13). In addition, CASC2/miR-21/PTEN axis modulates cisplatin sensitivity of CC (14). It was discovered that RGMB antisense RNA 1 (RGMB-AS1) showed oncogenic property in cancers such glioma (15), laryngeal squamous carcinoma (16), and lung adenocarcinoma (17). However, no report has linked RBMB-AS1 to CC yet.
The aim of this research was to inquire the biological role and possible mechanism of RGMB-AS1 in CC. We identified marked upregulation of RGMB-AS1 in CC tissues and cells and demonstrated that RGBM-AS1 elicited promoting function of in proliferation, apoptosis and invasion of CC cells via regulating microRNA-4428 (miR-4428)/PBX homeobox 1 (PBX1) axis.
Methods
Human tissue samples
All patients (n=32) were diagnosed with CC in the First Affiliated Hospital of Hebei North University Prior to this study, and these patients did not receive any other treatment. Fresh CC tissues and corresponding para-tumor tissues were gained from abovementioned participants hospitalized from May 2013 to December 2018. Fresh tissues were frozen in liquid nitrogen immediately after resection and stored at –80 °C. Written informed consent was signed by every patient and this experiment gained permission from ethics committee of the First Affiliated Hospital of Hebei North University (approval number: 1809036).
Cell culture
Human cervical epithelial cell (H8) and CC cells (HeLa, C-33A, SiHa and CaSki) were bought from Chinese Academy of Sciences (Beijing, China). A RPMI-1640 medium (Invitrogen, Carlsbad, USA) containing 10% FBS (Invitrogen) plus 1% penicillin/streptomycin (Sigma-Aldrich, Milan, Italy) was applied to culture above cells. A moist environment of 5% CO2 at 37 °C was required for culturing. Besides, medium was replaced every 3 days.
Cell transfection
Specific shRNAs against RGMB-AS1 (sh-RGMB-AS1#1 and sh-RGMB-AS1#2) with corresponding negative control (sh-NC), and the pcDNA3.1 vector targeting RGMB-AS1 or PBX1 and with negative control vector, were gained from Genechem (Shanghai, China). Moreover, miR-4428 mimics and NC mimics were gained from GenePharma (Shanghai, China). Above plasmids were separately transfected into SiHa or CaSki cells utilizing Lipofectamine 3000 (Invitrogen, Carlsbad, USA).
Quantitative real-time PCR (qRT-PCR)
Isolation of cellular RNA was finished via TRIZOL (Invitrogen, Carlsbad, USA) and cDNA was synthesized by iScriptTM Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA). Samples were sequentially analyzed with SYBR Green Master Mix (Vazyme, Nanjing, China) on ABI 7500 Realtime PCR system (Applied Biosystems, Foster City, CA, USA). Fold changes of target genes were calculated as per 2–ΔΔCt method and GAPDH or U6 served as the endogenous calibrator control.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was explored with CCK-8 (Dojindo, Kumamoto, Japan). Transfected SiHa or CaSki cells were inoculated into 96-well plates, and cultured overnight. Upon this, before detection, each well was supplemented with 10 µL CCK-8 solutions. Values at 450 nm were examined through the Epoch Microplate Spectrophotometer (Bio Tek, Winooski, VT, USA) upon culture for 0, 24, 48, 72 and 96 h, respectively.
Western blot
Total protein was obtained from transfected SiHa or CaSki cells which were lysed by radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Jiangsu, China). Subsequently, protein was quantified by using the BCA assay kit (Beyotime). The dodecyl sulfate, sodium salt (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed on proteins from lysates which were then moved to polyvinylidene fluoride membranes (PVDF; Millipore, Bedford, MA, USA). Membranes were cultured with primary antibodies overnight at 4 °C, and antibodies used were as follows: anti-Bcl-2 (ab32124, Abcam, Cambridge, USA), anti-Bax (ab32503, Abcam), anti-MMP2 (ab215986, Abcam), anti-MMP9 (ab219372, Abcam), anti-PBX1 (ab97994, Abcam) and anti-GAPDH (ab97994, Abcam). Then, secondary antibodies were added for cultivation for 1 h in dark room. Finally, the proteins were detected by an Enhanced Chemiluminescence Detection Kit (Millipore, Bedford, MA, USA).
Terminal dexynucleotidyl transferase (TdT)-mediated dUTP nick end labeling assay
The apoptosis of transfected SiHa or CaSki cells was assessed via a TUNEL Apoptosis Kit (Invitrogen, Carlsbad, USA). Cells were dyed with DAPI (Koritai, Beijing, China). Then, cells were observed and images were captured by using a fluorescence microscopy (Olympus, Tokyo, Japan).
5-ethynyl-2’-deoxyuridine (EdU) assay
Transfected CaSki or SiHa cells were seeded into fresh 96-well plates for cultivation. After incubation with EdU (Sigma-Aldrich, St. Louis, MO, USA) for 2 h, the cells were fixed by using 4% PFA (Solarbio, Beijing, China) and dyed in Apollo Dye Solution (RiboBio, Guangdong, China). Nucleic acid was stained with Hoechst 33342 stain (Invitrogen, Carlsbad, USA). Images were captured under an inverted fluorescence microscope (Carl Zeiss, Jena, Germany) and the proportion of EdU-positive cells was determined.
Cell invasion assay
Cell invasion was studied utilizing the 24-well Transwell system (Corning Costar, Cambridge, MA, USA). Transfected SiHa or CaSki cells were trypsinized and suspended in serum-free medium and subsequently added to the upper chamber of the Transwell system with Matrigel (BD Biosciences, San Jose, CA, USA). A 0.6 mL medium with additional 10% FBS was plated to the lower chamber. Cells were cultured for 24 h for invasion test. Invasive cells were fixed and stained utilizing crystal violet (Richard-Allan Scientific, San Diego, CA, USA), followed by washed twice using PBS (Sigma-Aldrich). Stained cells were eventually observed by an inverted microscope (Olympus Co., Tokyo, Japan).
Subcellular fractionation
For separating nuclear and cytoplasmic fraction, RNAs from SiHa or CaSki cells were isolated with the PARIS kit (Thermo Fisher Scientific, Waltham, MA, USA). The RNA expression level of RGMB-AS1 in nuclear and cytoplasmic fractions was explored using qRT-PCR.
Luciferase reporter assay
RGMB-AS1-WT/Mut or PBX1-WT/Mut was sub-cloned into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) so as to generate pmirGLO-RGMB-AS1-WT/Mut or pmirGLO-PBX1-WT/Mut. The pmirGLO-RGMB-AS1-WT/Mut was co-transfected into SiHa or CaSki cells with miR-4428 mimics or NC mimics. The pmirGLO-PBX1-WT/Mut was co-transfected into SiHa or CaSki cells with miR-4428 mimics or miR-4428 mimics + pcDNA3.1/RGMB-AS1 or NC mimics. At 48 h post-transfection, dual luciferase reporter assay system (Promega, Madison, WI, USA) was applied to examine luciferase activities.
RNA pull-down
SiHa or CaSki cells were lysed using lysis buffer and incubated with Bio-miR-4428-WT/Mut or Bio-NC, followed by incubation with streptavidin-coupled magnetic beads (Invitrogen). RGMB-AS1 and PBX1 levels were individually studied by qRT-PCR.
RNA immunoprecipitation (RIP)
RIP analysis was conducted in SiHa or CaSki cells using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Cells were lysed in RIP buffer. The resulting cell extraction was incubated with ProteinA/G magnetic beads bounded to anti-Ago2 (Abcam, Cambridge, UK) or anti-IgG (Abcam). Enrichment levels of RGMB-AS1, miR-4428 and PBX1 were individually measured with qRT-PCR.
Statistical analysis
Data were expressed as means ± SD. Statistical analysis was conducted using the SPSS (IBM, Armonk, NY, USA). Significance of the variance between two or several groups was evaluated by Student’s t-test or one-way ANOVA. Correlation among RGMB-AS1, miR-4428 and PBX1 was assessed by Pearson’s correlation analysis. P<0.05 was regarded as statistically significant in general. The experiments were conducted for at least thrice.
Results
RGMB-AS1 is overexpressed in HCC and facilitates CC cell proliferation, invasion and restrained cell apoptosis
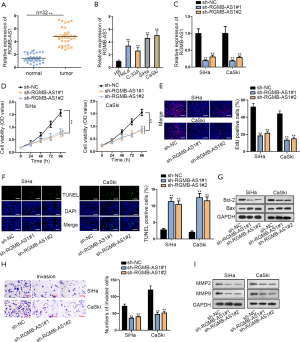
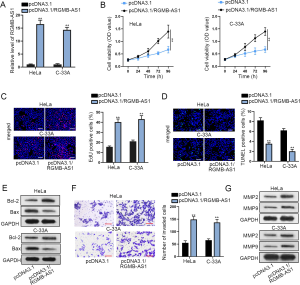
To explore the expression pattern of RGMB-AS1 in CC tissues and cell lines (HeLa, C-33A, SiHa, CaSki), qRT-PCR analysis was conducted. The adjacent non-tumor tissues and cervical epithelial cell (H8) were utilized as negative controls, separately. As a result, RGMB-AS1 expression was dramatically elevated in CC tissues (Figure 1A). Consistently, the high RGMB-AS1 level was found in CC cell lines (Figure 1B). To estimate the role of RGMB-AS1 in CC cells, RGMB-AS1 was knocked down by sh-RGMB-AS1#1 and sh-RGMB-AS1#2 (Figure 1C). Then, CCK-8 data and EdU-labeling results indicated that SiHa and CaSki cell proliferation was remarkably reduced by RGMB-AS1 knockdown of (Figure 1D,E). Later, the impact of RGMB-AS1 depletion on cell apoptosis was evaluated. Shown by the result of TUNEL assay, RGMB-AS1 deficiency significantly encouraged apoptosis of SiHa and CaSki cells (Figure 1F). The reduced Bcl-2 and increased Bax protein levels upon RGMB-AS1 silence were observed with Western blot assay, further verifying that cell apoptosis was promoted by RGMB-AS1 knockdown (Figure 1G). Moreover, transwell assay depicted decreased invasion ability in SiHa and CaSki cells (Figure 1H). Lastly, the effect of RGMB-AS1 on the proteins that related to metastasis were assessed. The results showed that silenced RGMB-AS1 considerably restrained the levels of MMP2 and MMP9 proteins (Figure 1I). Additionally, RGBM-AS1 was overexpressed by pcDNA3.1/RGMB-AS1 in HeLa and C-33A cells which expressed relatively low RGMB-AS1 level (Figure S1A). Then, we discovered that overexpressing RGMB-AS1 drive proliferation and depressed apoptosis in 2 cell lines (Figure S1B,C,D). Accordingly, Bcl-2 level upregulated and Bax level de-regulated under RGMB-AS2 overexpression (Figure S1E). Invasion of HeLa and C-33A cells was weakened upon RGMB-AS1 overexpression (Figure S1F), and MMP2 and MMP9 levels declined with RGMB-AS1 overexpression as well (Figure S1G). Conclusively, RGMB-AS1 is overexpressed in HCC and facilitates CC cell proliferation, invasion and restrained cell apoptosis.
RGMB-AS1 was associated with miR-4428 by acting as a sponge
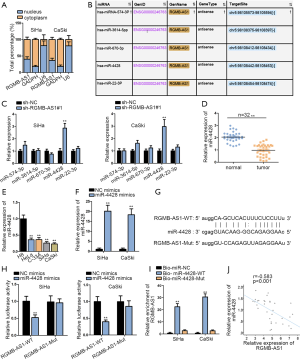
Mounting reports have suggested that lncRNAs and miRNAs were closely linked to regulate biological processes (18,19). To confirm whether RGMB-AS1 played regulatory role in CC via functioning as a ceRNA, its cellular localization was detected. As a result, RGMB-AS1 was more in cytoplasm than in nucleus (Figure 2A). Hence, we hypothesized that RGMB-AS1 might serve as a ceRNA in cytoplasm Thereafter, we applied StarBase tool (http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA&flag=target&clade=mammal&genome=human&assembly=hg19&miRNA=all&clipNum=1&deNum=0&panNum=0&target=RGMB-AS1) to search potential miRNAs for RGMB-AS1. The results manifested five miRNAs (miR-574-3p, miR-3614-5p, miR-670-3p, miR-4428 and miR-22-3p) that could interact with RGMB-AS1 (Figure 2B). As illustrated in Figure 2C, only miR-4428 expression was notably increased in CC cells transfected with sh-RGMB-AS1#1. Therefore, miR-4428 was selected for the following investigation. To confirm the relationship between miR-4428 and CC, miR-4428 expression was measured with qRT-PCR analysis. The result displayed that miR-4428 expression was obviously under-expressed in CC tissues and cells versus the corresponding normal tissues and cervical epithelium (Figure 2D,E). Then, we verified the elevated miR-4428 expression after transfecting miR-4428 mimics in 2 CC cell lines (Figure 2F). Based on StarBase prediction, a miR-4428 binding site in RGMB-AS1 was recognized, and we mutated this site for further detection (Figure 2G). Consequently, the luciferase activity for RGMB-AS1-WT declined evidently under miR-4428 overexpression, but that of RGMB-AS1-Mut was unchanged (Figure 2H). RNA pull-down assay uncovered that RGMB-AS1 was pulled down by miR-4428-WT rather than miR-4428-Mut biotinylated probe (Figure 2I). At last, the negative relevance between RGMB-AS1 and miR-4428 was recognized in CC tissues (Figure 2J). Taken together, RGMB-AS1 interacted with miR-4428 as a sponge.
RGMB-AS1 upregulated PBX1 by sponging miR-4428
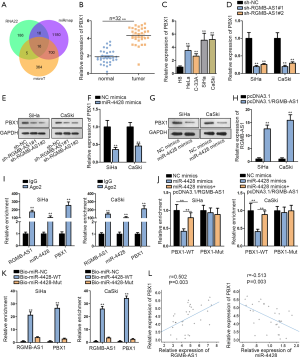
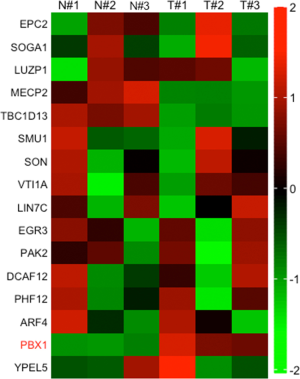
To support the hypothesis of ceRNA mechanism, we subsequently searched the downstream mRNAs for miR-4428 using StarBase with the setting of 3 prediction programs (RNA22, miRmap and microT). The Venn diagram showed that there are 16 potential mRNAs for miR-4428 (Figure 3A). qRT-PCR analysis showed that among 16 candidate genes, PBX1 exhibited the most significant upregulation in 3 CC specimens versus corresponding para-tumor normal ones (Figure S2). PBX1 was commonly reported as oncogene (20-22), so we speculated PBX1 as the target for miR-4428 in CC. qRT-PCR analysis confirmed the upregulated PBX1 in 32 CC tissues and 4 cell lines (Figure 3B,C). Then, effects of RGMB-AS1 and miR-4428 on PBM1 were measured. As presented in Figure 3D,E, the levels of PBX1 mRNA and protein were diminished with the transfection of sh-RGMB-AS1#1/2. Besides, miR-4428 mimics observably attenuated PBX1 mRNA and protein levels (Figure 3F,G). Then, the overexpression of RGMB-AS1 by pcDNA3.1/RGMB-AS1 in CC cells was confirmed by qRT-PCR (Figure 3H). Moreover, as shown in Figure 3I, RIP assay demonstrated that RGMB-AS1, miR-4428 and PBX1 could be detected in Ago2 immunoprecipitates rather than IgG (Figure 3I). In addition, luciferase reporter assay confirmed that the decrease of PBX1-WT luciferase activity caused by miR-4428 mimics was restored by RGMB-AS1 overexpression, but no evident difference was discovered in PBX1-Mut (Figure 3J). RNA pull-down assay further validated that both RGMB-AS1 and PBX1 were pulled down by miR-4428-WT, rather than miR-4428-Mut (Figure 3K). Furthermore, PBX1 expression was positively associate with RGMB-AS1 and negatively related to miR-4428 in CC specimens (Figure 3L). Jointly, RGMB-AS1 regulates PBX1 by sponging miR-4428.
RGMB-AS1 enhances CC progression through regulating PBX1
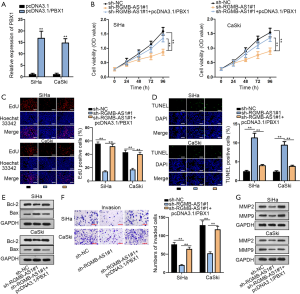
Finally, we tested whether PBX1 was a target for RGMB-AS1 to regulate CC cellular activities. We first used pcDNA3.1/PBX1 to increase the expression of PBX1 so as to unfold rescue experiments (Figure 4A). Later, we showed that the proliferative ability of CC cells was restrained by the silence of RGMB-AS1 and was recovered by PBX1 overexpression (Figure 4B,C). Besides, TUNEL staining suggested that the promoting effect of RGMB-AS1 silence on CC cell apoptosis was offset by transfecting pcDNA3.1/PBX1 through TUNEL (Figure 4D). Decreased Bcl-2 and increased Bax in CC cells with RGMB-AS1 silence were reversed by PBX1 overexpression (Figure 4E). According to transwell assay, overexpressing PBX1 countervailed the repressive impact of RGMB-AS1 depletion on CC cell migration (Figure 4F). Additionally, silenced RGMB-AS1 lessened the levels of MMP2 and MMP9, while the transfection of pcDNA3.1/PBX1 reversed the effect of silenced RGMB-AS1 (Figure 4G). In brief, RGMB-AS1 enhances CC progression through regulating PBX1.
Discussion
It was proposed that lncRNAs could function as regulators in the biological processes, including cancers. Recent studies have revealed that lncRNAs might act as sponges for miRNAs in cancer progression (23). Several lncRNA-miRNA axes are revealed in cancers, such as MALAT1-miR-218 axis in choriocarcinoma (24), NEAT1-miR-181a-5p in lung cancer (25), and PVT1-miR-128-3p axis in CC (26). Data form previous volumes supported that RGMB-AS1 is activated by E2F1 and enhances papillary thyroid carcinoma cell proliferation and invasion (27). RGMB-AS1 is also associated with clinical stage in hepatocellular carcinoma (28). RGMB-AS1 indicates an unfavorable prognosis in lung adenocarcinoma and regulates its progression (17). Hence, we assumed that RGMB-AS1 might participate in CC. Expectedly, our study first suggested that RGMB-AS1 was significant overexpressed in CC tissues and cells. RGMB-AS1 knockdown lessened proliferation and invasion, and increased apoptosis, whereas its overexpression had opposite effects. These data consistently implied the oncogenic property of RGMB-AS1 in CC.
MiRNAs are a kind of short RNAs with 21–25 nucleotides (29). MiRNA serves as tumor promoters or tumor suppressors in tumorigenesis and development of cancers (30,31). For instance, miRNA-518 represses cell growth and induces apoptosis in gastric cancer via targeting MDM2. MiR-183 is overexpressed in glioblastoma and downregulates LRIG1 expression to promote glioblastoma radioresistance (32). MiRNA-423 promotes hepatocellular carcinoma cell invasion via regulating BRMS1 (33). A variety of miRNAs was reported to relate to CC progression. For example, miRNA-433 represses CC tumor size and distant metastasis by regulating the AKT/β-catenin signaling pathway and targeting to metadherin (34). MicroRNA-150 targets to PDCD4 in CC and boosts cell proliferation, migration and invasion (35). MiR-214 enhances the drug sensitivity of CC via regulating FOXM1 (36). MiR-4428 is first identified as a downregulated miRNA in CC tissues and cells. Further, we first predicted and demonstrated that RGMB-AS1 directly combine with miR-4428 via putative MRE. Moreover, we first exhibited that RGMB-AS1 expression was negatively correlated with miR-4428 expression in CC. Thus, we provided first data to link RGMB-AS1 to miR-4428 in CC.
Further, we found that PBX1 was a target for miR-4428 and upregulated overtly in CC. As formerly reported, PBX1 promotes the proliferation of clear cell renal carcinoma by JAK2/STAT3 signaling (20). PBX1 is upregulated in breast cancer and plays essential role in the regulation of breast cancer development (21). PBX1 serves as a stem cell reprogramming factor and modulates the chemoresistance of ovarian cancer (22). However, the role and mechanism of PBX1 in CC is first explained by our study. We showed that both RGMB-AS1 knockdown and miR-4428 overexpression could decline the expression of PBX1. Importantly, we found that RGMB-AS1 regulates PBX1 expression by sponging miR-4428. Moreover, we delineated the positive relevance between RGMB-AS1 and PBX1 and the negative relevance between miR-4428 and PBX1. According to rescue experiments, PBX1 overexpression restored RGMB-AS1 depletion-mediated repressing effect on CC progression, indicating that PBX1 was a target for RGMB-AS1 to regulate CC.
Conclusively, RGMB-AS1 contributes to CC progression through acting as a ceRNA, and RGMB-AS1/miR-4428/PBX1 axis could offer an inspiring thought for the therapy in CC patients.
Acknowledgments
We appreciate all the participants who provide supports in this study.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.04.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was signed by every patient and this experiment gained permission from ethics committee of the First Affiliated Hospital of Hebei North University (approval number: 1809036).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Musselwhite LW, Oliveira CM, Kwaramba T, et al. Racial/ethnic disparities in cervical cancer screening and outcomes. Acta Cytol 2016;60:518-26. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Wentzensen N, Schiffman M. Accelerating cervical cancer control and prevention. Lancet Public Health 2018;3:e6-7. [Crossref] [PubMed]
- Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 2018;33:706-20.e9. [Crossref] [PubMed]
- Li J, Liu C. Coding or noncoding, the converging concepts of RNAs. Front Genet 2019;10:496. [Crossref] [PubMed]
- Zhang H, Bai M, Zeng A, et al. LncRNA HOXD-AS1 promotes melanoma cell proliferation and invasion by suppressing RUNX3 expression. Am J Cancer Res 2017;7:2526-35. [PubMed]
- Zhang LM, Wang P, Liu XM, et al. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res 2017;9:5461-72. [PubMed]
- Li Y, Wen X, Wang L, et al. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol 2017;26:527-34. [Crossref] [PubMed]
- Furió-Tarí P, Tarazona S, Gabaldón T, et al. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res 2016;44:W176-80. [Crossref] [PubMed]
- Qu J, Li M, Zhong W, et al. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med 2015;8:17110-6. [PubMed]
- He X, Guo S, Wang Y, et al. Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.). Physiol Plant 2020;168:736-54. [Crossref] [PubMed]
- Shi C, Yang Y, Yu J, et al. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res 2017;7:2157-68. [PubMed]
- Zhu H, Zeng Y, Zhou CC, et al. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys 2018;637:1-8. [Crossref] [PubMed]
- Feng Y, Zou W, Hu C, et al. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys 2017;623-624:20-30. [Crossref] [PubMed]
- Pan B, Zhao M, Wang N, et al. LncRNA RGMB-AS1 promotes glioma growth and invasion through miR-1200/HOXB2 axis. Onco Targets Ther 2019;12:10107-14. [Crossref] [PubMed]
- Xu Z, Xi K. LncRNA RGMB-AS1 promotes laryngeal squamous cell carcinoma cells progression via sponging miR-22/NLRP3 axis. Biomed Pharmacother 2019;118:109222. [Crossref] [PubMed]
- Li P, Zhang G, Li J, et al. Long noncoding RNA RGMB-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS One 2016;11:e0150790. [Crossref] [PubMed]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018;109:2093-100. [Crossref] [PubMed]
- Wei W, Liu Y, Lu Y, et al. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem 2017;118:3349-58. [Crossref] [PubMed]
- Wei X, Yu L, Li Y. PBX1 promotes the cell proliferation via JAK2/STAT3 signaling in clear cell renal carcinoma. Biochem Biophys Res Commun 2018;500:650-7. [Crossref] [PubMed]
- Wang J, Shidfar A, Ivancic D, et al. Overexpression of lipid metabolism genes and PBX1 in the contralateral breasts of women with estrogen receptor-negative breast cancer. Int J Cancer 2017;140:2484-97. [Crossref] [PubMed]
- Jung JG, Shih IM, Park JT, et al. Ovarian cancer chemoresistance relies on the stem cell reprogramming factor PBX1. Cancer Res 2016;76:6351-61. [Crossref] [PubMed]
- Xu W, Yu S, Xiong J, et al. CeRNA regulatory network-based analysis to study the roles of noncoding RNAs in the pathogenesis of intrahepatic cholangiocellular carcinoma. Aging (Albany NY) 2020;12:1047-86. [Crossref] [PubMed]
- Shi D, Zhang Y, Lu R, et al. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother 2018;97:543-50. [Crossref] [PubMed]
- Li S, Yang J, Xia Y, et al. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res 2018;26:289-96. [Crossref] [PubMed]
- Gao YL, Zhao ZS, Zhang MY, et al. Long noncoding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR-424. Oncol Res 2017;25:1391-8. [Crossref] [PubMed]
- Zhang Z, Li SY, Zhang LB. LncRNA RGMB-AS1 is activated by E2F1 and promotes cell proliferation and invasion in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci 2018;22:1979-86. [PubMed]
- Sheng N, Li Y, Qian R, et al. The clinical significance and biological function of lncRNA RGMB-AS1 in hepatocellular carcinoma. Biomed Pharmacother 2018;98:577-84. [Crossref] [PubMed]
- Mohr S, Doebele C, Comoglio F, et al. Hoxa9 and Meis1 cooperatively induce addiction to Syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell 2017;31:549-62.e11. [Crossref] [PubMed]
- Lan H, Lu H, Wang X, et al. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int 2015;2015:125094.
- Wang Y, Wang B, Zhou H, et al. MicroRNA-384 inhibits the progression of papillary thyroid cancer by targeting PRKACB. Biomed Res Int 2020;2020:4983420.
- Fan H, Yuan R, Cheng S, et al. Overexpressed miR-183 promoted glioblastoma radioresistance via down-regulating LRIG1. Biomed Pharmacother 2018;97:1554-63. [Crossref] [PubMed]
- Sun X, Wang M, Liu H, et al. MicroRNA-423 enhances the invasiveness of hepatocellular carcinoma via regulation of BRMS1. Am J Transl Res 2017;9:5576-84. [PubMed]
- Liang C, Ding J, Yang Y, et al. MicroRNA-433 inhibits cervical cancer progression by directly targeting metadherin to regulate the AKT and β-catenin signalling pathways. Oncol Rep 2017;38:3639-49. [PubMed]
- Zhang Z, Wang J, Li J, et al. MicroRNA-150 promotes cell proliferation, migration, and invasion of cervical cancer through targeting PDCD4. Biomed Pharmacother 2018;97:511-7. [Crossref] [PubMed]
- Wang JM, Ju BH, Pan CJ, et al. MiR-214 inhibits cell migration, invasion and promotes the drug sensitivity in human cervical cancer by targeting FOXM1. Am J Transl Res 2017;9:3541-57. [PubMed]


