Construction of a prognostic model for lung adenocarcinoma based on bioinformatics analysis of metabolic genes
Introduction
Lung cancer is one of the most common malignancies and seriously threatens human life and quality of life. The morbidity and mortality of lung cancer ranks first among all cancers in the world, showing an increasing trend year by year (1). Lung cancer can be divided into small cell lung cancer and non-small cell lung cancer (NSCLC). NSCLC accounts for approximately 2/3 of the total cases of lung cancer. Lung adenocarcinoma (LUAD) is a major pathological subtype of NSCLC. The incidence of LUAD is higher than that of squamous cell lung cancer (2). Since the early onset of LUAD is relatively occult, most patients are in advanced stages with extensive surrounding tissue invasion and lymph node metastasis at the time of diagnosis. Patients often receive treatments that include surgery, radiotherapy, chemotherapy and targeted therapy; however, due to the lack of effective biomarkers for diagnosis and patient prognosis, most patients still have a high risk of tumor recurrence and spread, and only 15% of patients survive more than 5 years (3,4). Previous risk assessments and treatment plans for patients with LUAD were mostly based on risk factors such as tumor size, stage and lymph node metastasis (5). However, these clinicopathological risk factors cannot clearly distinguish high-risk patients and low-risk patients. Therefore, to reduce mortality in patients with LUAD, good prognostic indicators must be established to guide treatment and clinical management.
Depending on cellular functional requirements and metabolic capacity, cells can use various metabolic pathways for energy production and biosynthesis (6). As an important feature of tumors, changes in cell metabolism are causally related to tumorigenesis (7,8). As early as 1924, Otto Warburg discovered that compared with normal mature cells, tumor cells generate energy to meet the requirements of rapid growth by absorbing more glucose with higher efficiency. Even in aerobic conditions, tumor cells breakdown excess glucose mainly through glycolysis, which is accompanied by the production of a large amount of lactic acid. This is known as the Warburg effect (9). The extensive application of modern molecular biology technologies has greatly promoted the development of tumor metabolism research, and the connotation of the Warburg effect has been further expanded. At present, the Warburg effect is no longer limited to changes in glycolysis and the tricarboxylic acid cycle. The Warburg effect now also includes many metabolic pathways, such as fatty acid metabolism, glutamine metabolism, serine metabolism, one-carbon metabolism and choline metabolism (10). Abnormal tumor cell metabolism mainly manifests as glucose decomposition by aerobic glycolysis; reduced oxidative phosphorylation; enhanced pentose phosphate pathway activity, active glutamine catabolism, de novo fatty acid synthesis and active β-oxidation (11).
In previous studies, researchers found that to better adapt to the fluctuation of oxygen supply in the tumor growth environment, obtain raw materials required for growth and division, resist oxidative stress and trauma, thereby facilitating their own survival and metastasis, tumor cells must change their original metabolic model, i.e., the Warburg effect must be initiated (12). In fact, the metabolic changes in tumor cells are closely related to various stages of tumor occurrence and development. Overcoming telomere replication restrictions, reprogramming intracellular gene expression, resisting apoptosis, achieving immune escape and enhancing neovascularization all affect tumor cell metabolism to varying degrees (13). With the development of research techniques in cancer biology, the theory that metabolic abnormalities precede the occurrence and development of tumors has gradually been experimentally confirmed. Yun et al. found that glucose deficiency promoted the acquisition of mutations in KRAS and its signaling molecules in KRAS wild-type cells, indicating that abnormal cell metabolism can lead to proto-oncogene mutations (14), and Hu et al. used 13C-labeled pyruvate molecular imaging technologies in animals and found that metabolic changes in glycolysis preceded the formation and regression of c-Myc-induced tumors (15). In addition, Xu et al. also found that α-ketoglutarate exhibited competitive inhibition of various α-ketoglutarate-dependent dioxygenase activities to induce cancers (16). These experiments highlight the important role of abnormal cell metabolism in the occurrence and development of tumors and in the prognosis of patients with tumors, indicating that metabolic genes may serve as prognostic markers for lung cancers.
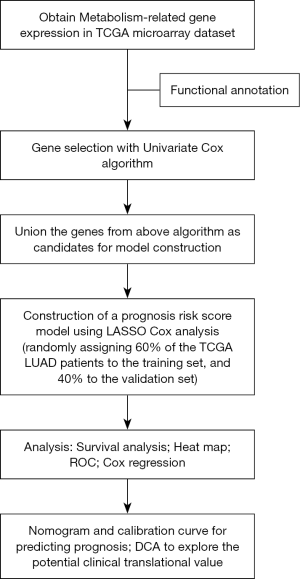
However, metabolism is a complex biological process involving hundreds of molecules. Therefore, compared to a single metabolic gene, a comprehensive model consisting of multiple metabolic genes can improve the accuracy of patient prognosis. Different from traditional prediction indicators composed of a single molecule, models composed of multiple genes have higher accuracy in predicting the prognosis of patients. Taking this into consideration, we used univariate Cox regression analysis to screen and identify prognosis-related metabolic genes from a training cohort consisting of 288 patients with LUAD. Then, the obtained genes were subjected to least absolute shrinkage and selection operator (LASSO) regression to establish an optimal risk model to calculate the risk score for each patient. According to the median value of the calculated risk scores, the patients were divided into a high-risk group and low-risk group. The prognostic value of the model was assessed through survival analysis and further validated in a validation cohort consisting of 189 patients with LUAD. Moreover, a nomogram combining clinical features with the prognostic model was established to predict individual survival probability. Finally, we find a representative hub gene and carry on experiments to confirm it.
Methods
LUAD database and the clinical information of the patients
Metabolic gene expression profiles from 535 LUAD cases and 59 nontumor tissue samples as well as the clinical information of 522 patients with LUAD were downloaded from the official The Cancer Genome Atlas (TCGA) website (https://gdc-portal.nci.nih.gov/). After excluding patients with incomplete clinical data and patients with survival times less than 30 days, this study included data from 477 patients for analysis. The relevant clinical data included overall survival (OS), age, sex, TNM staging and pathological grade. The 477 patients were randomly divided into two groups based on a 3:2 ratio; among them, 288 patients were included in the training cohort for constructing the prognostic model, while the remaining 189 patients belonged to the validation cohort to validate the prognostic ability of the model. Furthermore, the prognostic gene signature was verified in the independent LUAD cohorts (GSE37745) (17). Table 1 provides detailed information for the datasets used in this study.
Table 1
| Characteristics | Training cohort (n=288) | Verifying cohort (n=189) | TCGA total cohort (n=477) | GSE37745 (n=105) |
|---|---|---|---|---|
| Vital stat | ||||
| Alive | 186 | 119 | 305 | 29 |
| Dead | 102 | 70 | 172 | 76 |
| Age (years) | ||||
| <65 | 129 | 92 | 221 | 52 |
| ≥65 | 159 | 97 | 256 | 53 |
| Sex | ||||
| Female | 159 | 97 | 256 | 60 |
| Male | 129 | 92 | 221 | 45 |
| Stage | ||||
| I | 158 | 109 | 267 | 70 |
| II | 67 | 40 | 107 | 18 |
| III/IV | 63 | 40 | 103 | 17 |
LUAD, lung adenocarcinoma.
Acquisition and processing of the metabolic gene expression profiles of patients with LUAD
The keyword “metabolism” was inputted into the GENECARDS website (https://www.genecards.org/) to search metabolic genes. Correlation coefficients were used to indicate the degree of the correlation between genes and metabolism, ranging from 0 to 100; the higher the score was, the closer the correlation. Genes with a correlation coefficient greater than 10 were used as candidate metabolic genes. R software (Version 3.6.0; https://www.R-project.org) was used to retrieve the expression profiles of 1062 metabolic genes from TCGA. Subsequently, the R software package “Limma” (18) was used for log2 conversion of mRNA sequencing. The absolute values of log2FC >1 and adjusted P<0.05 were used as the threshold values for normalization and differential expression analysis. To explore the potential biological relevance of differentially expressed genes, the DAVID bioinformatics tool was used (https://david.ncifcrf) for Gene Ontology (GO) enrichment analysis of coexpressed genes. Using the entire human genome as the background, the KO-based annotation system bioinformatics tool (http://kobas.cbi.pku.edu.cn/) was applied for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of metabolic genes. Functional enrichment analysis results (P<0.05) were used to determine potential biological functions. Clustering and visualization of functionally similar GO and KEGG enrichment analyses were performed using the R software package “ggplot2”.
Construction of a predictive model of metabolic genes
Univariate Cox regression analysis was performed to evaluate the relationship between the expression of various differentially expressed genes and the OS of patients with LUAD. Considering the number of selected metabolic genes and their relationship with prognosis, genes with P<0.05 were identified as candidate genes. The candidate genes were further subjected to LASSO regression analysis. Lasso regression is a punishment strategy suitable for high-dimensional data that can prevent overfitting (19). In this study, we used 10-fold cross-validation to determine the value of λ, and we choose λ with the least biases of partial likelihood as the optimal λ. Then, we used the identified predictive genes to construct a risk prediction model based on gene expression, as shown below.
where N is the number of genes, Expi is the expression level of the gene, and Ci is the regression coefficient obtained from the LASSO regression analysis in the training cohort. The C-index was calculated to preliminarily assess the prediction accuracy of the model. Here, we propose that a C-index greater than 0.7 in the training cohort and the validation cohort indicates that the prognostic model is reliable and stable. The above method was implemented using GLIME and Hmisc in the R software package.
Analysis of protein-protein interaction (PPI) networks and find the hub gene
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/cgi/input.pl) provides experimental and predicted proteins which are interacted with each other. STRING analysis was executed to form a PPI network, with the criterion of a combined score of >0.4. Besides, the 16 metabolic genes in the prognostic model were used as queries in the STRING database and the resultant PPI network was subsequently visualized by Cytoscape software. The top one hub gene was found by Cytoscape software with CytoHubba which is a plug-in in Cytoscape software.
Expression level of the hub gene (TYMS) by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry
A total of 50 patient data with LUAD were collected (30 females and 20 males; composite life was 64.55 years; range, 36 to 82 years), with both LUAD and adjacent lung tissue obtained through surgery and provided by Second Affiliated Hospital of the Guangxi Medical University and Chengdu Medical College, from May 2013 to July 2019. The experiments were approved by the Ethical Committee of the two hospitals and written informed consent was signed by each participant. Total RNA was isolated from the lung tissue using the Trizol reagent (ThermoFisher, the United States) according to the kit instructions, and a PCR amplification kit (ABI, Life, Technologies, the United States). Synthesis of cDNA was implement using the Superscript first strand cDNA synthesis kit according to the manufacturer’s protocol. RT-qPCR was carried out using ABI 7500 Prep-Station and the SYBR®-Green PCR Master Mix. PCR was performed first with a 10 minutes hot start, followed with 95°C for 15 seconds, and 60 °C for 30 seconds for 40 cycles. The specific primers used were the following: TYMS forward:5'-GGCACCCTGTCGGTATTCG-3', adverse:5'-CCCTTCCAGAACACACGTT-3' and β-actin (internal control) forward, 5'-CATCCTCACCCTFAAGTA-3', and reverse, 5'-ACACGCAGCTCATTGTAG-3'. The results were normalized to β-actin expression and calculated by the 2−ΔCq method. All RT-qPCR reactions and test were executed at least three times, repeatedly.
In the present study, TYMS protein expression was detected by immunohistochemistry (IHC) with TYMS antibody (Abcam, the United States). SPlink Detection kits (Biotin-Streptavidin HRP Detection Systems; SP-9000; Beijing, China) was applied to the IHC.
Sections were blocked with 10% goat serum for 20 minutes at indoor temperature. Next, the primary antibody was added and incubated for one hour at indoor temperature.
Additionally, 100 µL of the secondary antibody (Goat Anti-Rabbit Immunoglobulin G) which is also included in the SPlink Detection kits was added for incubation that lasting 15 minutes. All the experimental procedures must follow the kit instructions. Five random images were captured with a light microscope. The percentage of positive TYMS staining was evaluated by 0−4 scores, which suggested to ≤10, >10 to ≤25, 25 to ≤50, 50 to ≤75 and 75 to ≤100%, respectively. The weak, moderate and strong intensities of TYMS staining were scored using 1, 2 and 3, respectively. The scoring criteria were examined by two independent pathologists. if the score was >2, positive TYMS staining would be confirmed; if the score was ≤2 negative TYMS staining would be confirmed.
Statistical analysis
After modeling 10 million times, a prognostic model consisting of 16 metabolic genes was successfully established. The model was used to calculate a risk score for each patient with LUAD. Based on the median risk score, the included patients were divided into a high-risk group and a low-risk group. Next, we performed time-dependent ROC curve analysis and calculated the area under the curve (AUC) with different survival times to measure the prediction accuracy of the established model. To further evaluate whether this model has advantages over other commonly used clinical parameters and whether this model is worthy of clinical application, we used decision curve analysis (DCA) to evaluate the prognostic model. The decision curve was plotted using the R software package “Decision Curve”. Next, we performed multivariate Cox regression analysis based on relevant clinical information such as age, sex, and pathological staging to verify whether the predictive power of the constructed model was independent of other clinical factors and whether the model could become an independent predictive factor. Multivariate regression analysis was performed to calculate regression coefficients and to plot a nomogram. The bootstrap method was used to evaluate the performance of the nomogram using the C-index. A calibration curve was used for visualization. In all analyses, P<0.05 was considered statistically significant. Statistical analysis was performed using SPSS 20.0 software, and GraphPad Prism 8.0 was used to plot the results of paired T test.
Results
Patient characteristics
The flowchart for the study is shown in Figure 1. Table 1 summarizes the common clinical characteristics of the patients. A total of 477 patients with LUAD were included in this study, including 288 in the training cohort and 189 in the validation cohort.
Differentially expressed metabolic genes
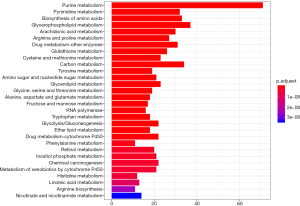
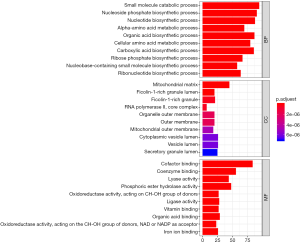
A total of 1,062 metabolic genes were obtained from the website GENECARDS. After differential expression analysis, we obtained 455 differentially expressed genes, 125 of which were upregulated and 330 of which were downregulated [Figures 2,3 (volcano plot and heat map)]. Table 2 lists the top ten differentially expressed genes. We further performed a series of GO enrichment analyses and KEGG pathway analyses to further study the biological functions of these metabolic genes in the development and progression of LUAD. GOplot analysis showed that these genes are all related to metabolism and biosynthesis in biological processes, as shown in Figure 4. In terms of cellular components, these genes are involved in the mitochondrial outer membrane and RNA core complex, as shown in Figure 4. Regarding molecular function, these genes also play an indispensable role in coenzyme binding, as shown in Figure 4. In addition, KEGG pathway enrichment analysis indicated that these genes are mainly enriched in purine metabolism and related pathways, as shown in Figure 5.
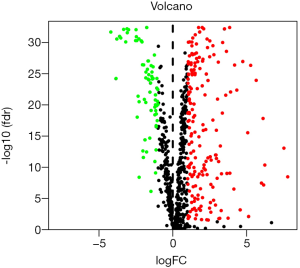
Table 2
| Gene symbol | logFC | P value | fdr | Regulation |
|---|---|---|---|---|
| CA4 | −4.19382 | 3.12E-34 | 2.05E-32 | Down |
| ALAS2 | −3.84285 | 7.46E-26 | 5.92E-25 | Down |
| GPD1 | −3.78296 | 7.28E-33 | 2.72E-31 | Down |
| CYP1A2 | −3.59665 | 2.04E-33 | 1.04E-31 | Down |
| OTC | −3.52038 | 4.27E-33 | 1.90E-31 | Down |
| INMT | −3.33088 | 8.52E-35 | 8.11E-33 | Down |
| ADCY8 | −3.18507 | 2.99E-32 | 8.12E-31 | Down |
| FMO2 | −3.09618 | 5.14E-35 | 6.52E-33 | Down |
| ADH1B | −3.07807 | 4.72E-34 | 2.77E-32 | Down |
| ACADL | −2.98845 | 1.16E-34 | 8.82E-33 | Down |
| AOC1 | 5.078884 | 9.26E-17 | 2.92E-16 | Up |
| AKR1C4 | 5.140016 | 0.006155 | 0.007552 | Up |
| ITPKA | 5.280638 | 3.94E-28 | 4.62E-27 | Up |
| CA9 | 5.652221 | 1.57E-25 | 1.16E-24 | Up |
| GPX2 | 6.013781 | 1.91E-08 | 3.41E-08 | Up |
| PLA2G2F | 6.121566 | 3.79E-19 | 1.48E-18 | Up |
| UGT2B15 | 6.158543 | 3.85E-08 | 6.78E-08 | Up |
| PAH | 6.237651 | 1.97E-11 | 4.31E-11 | Up |
| AKR1B10 | 7.530102 | 3.24E-14 | 8.50E-14 | Up |
| UGT2B11 | 7.79052 | 1.73E-09 | 3.28E-09 | Up |
The construction and validation of a prognostic model of LUAD using the training cohort
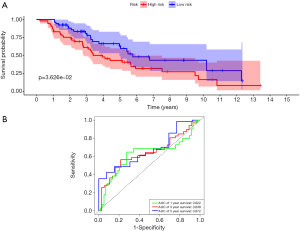
First, univariate Cox regression of the training cohort was performed to analyze the 455 differentially expressed genes. A total of 59 metabolic genes were selected as candidate genes associated with prognosis (P<0.05). To avoid overfitting, these candidate genes were subjected to LASSO regression analysis. After the construction of the model (50,000 times), we successfully extracted 16 key metabolic genes and constructed a reliable and stable risk scoring model based on the regression coefficients generated by LASSO regression analysis, as shown in Figure 6A,B and Table 3. The C-index of the model was 0.815 and 0.794 for the training cohort and validation cohort, respectively. Based on the expression levels of the 16 genes, a risk score was assigned to each patient, and then the patients were divided into a high-risk group (136 cases) and a low-risk group (152 cases) using the median risk score of 2.764. Kaplan-Meier survival analysis showed that the median survival time was 1.27 years in the high-risk group, which was significantly lower than that in the low-risk group (1.92 years) (P=2.183E-5) (Figure 7A). The 3- and 5-year survival rates for the high-risk group were 41.2% and 29.5%, respectively. The 3- and 5-year survival rates for the low-risk group were 73.7% and 43.2%, respectively. To further validate the applicability and stability of the model, we performed the same analysis using the validation cohort and found that the median risk score was 3.125 for the 189 patients with LUAD. According to this median score, the 189 patients were also divided into a high-risk group, with 102 patients, and a low-risk group, with 87 patients. Kaplan-Meier survival analysis showed that the OS of these two groups was significantly different. Specifically, the median survival time of the patients in the high-risk group was 1.51 years, which was significantly lower than that in the low-risk group (2.01 years) (P=5.83E-5). The 3- and 5-year survival rates for the high-risk group were 39.6% and 20.5%, respectively, and the 3- and 5-year survival rates for the low-risk group were 74.6% and 44.6%, respectively, as shown in Figure 7B. The time-dependent ROC curve plotted using the prognostic model consisting of 16 metabolic genes predicted AUC values of 0.732 and 0.69 for the 3- and 5-year survival rates, respectively, for the training cohort, indicating that the prognostic model had excellent reliability in predicting survival. The ROC curve is shown in Figure 8A. The predicted AUC values for the 3- and 5-year survival rates were 0.703 and 0.703 respectively, in the validation cohort, which was basically consistent with the training cohort, as shown in Figure 8B. Figure 9A,B,C show the risk score distribution, survival status, and heat map of the expression profiles of 16 metabolic genes of 288 patients with LUAD in the training cohort. The ALDOA, LDHA, and PTGES genes showed high expression in the high-risk group, while these genes showed low expression in the low-risk group. The risk score distribution, survival status and gene expression profile heat map of the validation cohort were consistent with those of the training cohort (Figure 9D,E,F). Univariate analysis was performed on the models consisting of 16 metabolic-related genes in the training and validation cohorts. The results suggested that this model was closely related to the survival of patients with LUAD (training cohort: HR =2.34, 95% CI: 1.64–3.26, P<0.05; validation cohort: HR =2.58, 95% CI: 1.86–3.83, P<0.05). Table 4 provides more detailed results.
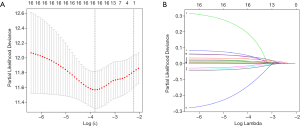
Table 3
| No. | Gene symbol | Full name | Function | Coefficient of risk |
|---|---|---|---|---|
| 1 | TYMS | Thymidylate Synthetase | Fluoropyrimidine activity and metabolism of nucleotides | 0.012000842 |
| 2 | GNPNAT1 | Glucosamine-Phosphate N-Acetyltransferase 1 | Synthesis of substrates in N-glycan biosynthesis | 0.02509742 |
| 3 | LPGAT1 | Lysophosphatidylglycerol Acyltransferase 1 | Glycerophospholipid biosynthesis | 0.010592199 |
| 4 | INPP4B | Inositol Polyphosphate-4-Phosphatase Type II B | Inositol phosphate metabolism | 0.029983089 |
| 5 | MAOB | Monoamine Oxidase B | Super pathway of tryptophan utilization | −0.013252539 |
| 6 | MTHFD2 | Methenyltetrahydrofolate Cyclohydrolase | Nucleotide metabolism and selenium micronutrient network | −0.005686523 |
| 7 | ADCY9 | Adenylate Cyclase 9 | Adenylate cyclase inhibition | −0.020756554 |
| 8 | PTGIS | Prostaglandin I2 Synthase | Prostaglandin 2 biosynthesis | 0.008160885 |
| 9 | ALOX12B | Arachidonate 12-Lipoxygenase, 12R Type | Arachidonic acid metabolism and prostaglandin 2 biosynthesis | 0.110691209 |
| 10 | GSTA3 | Glutathione S-Transferase Alpha 3 | Glutathione metabolism and platinum drug resistance | −0.08312793 |
| 11 | LDHA | Lactate Dehydrogenase A | Pyruvate metabolism and citric acid (TCA) cycle | 0.00196183 |
| 12 | UCK2 | Uridine-Cytidine Kinase 2 | Fluoropyrimidine activity and metabolism of nucleotides | 0.027401706 |
| 13 | ENTPD2 | Ectonucleoside Triphosphate Diphosphohydrolase 2 | Metabolism of nucleotides | 0.035619134 |
| 14 | PTG level | Prostaglandin E Synthase | Arachidonic acid metabolism | 0.001613886 |
| 15 | NT5E | 5'-Nucleotidase Ecto | Metabolism of nucleotides and NAD metabolism | 0.003867883 |
| 16 | ALDOA | Fructose-Bisphosphate Aldolase A | Innate immune system and carbon metabolism | 0.000733303 |
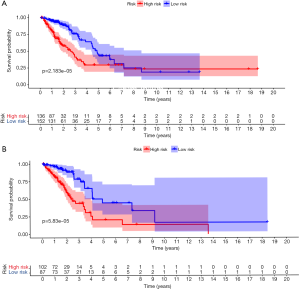
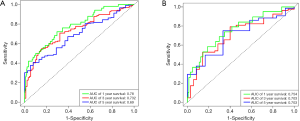
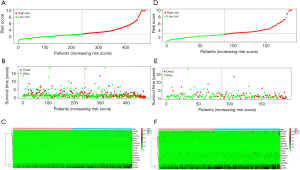
Table 4
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |||
| (I) Training cohort (n=288) | ||||||||
| Risk score for 16 metabolism-related genes (high/low risk) | 2.34 | 1.64–3.26 | <0.001 | 2.44 | 1.58–3.74 | <0.001 | ||
| Age (≥65/<65 years) | 1.24 | 0.82–2.20 | 0.19 | 1.11 | 0.89–1.73 | 0.87 | ||
| Sex (female/male) | 0.99 | 0.52–1.32 | 0.21 | 1.03 | 0.69–1.52 | 0.91 | ||
| Stage | ||||||||
| II vs. I | 1.72 | 1.11–2.92 | 0.04 | 1.74 | 1.21–3.24 | 0.03 | ||
| III/IV vs. I | 2.74 | 1.87–4.45 | <0.001 | 2.53 | 1.63–4.83 | <0.001 | ||
| (II) Verification cohort (n=189) | ||||||||
| Risk score for 16 metabolism-related genes (high/low risk) | 2.58 | 1.86–3.83 | <0.001 | 2.15 | 1.52–2.70 | 0.001 | ||
| Age (≥65/<65 years) | 1.20 | 0.88–1.67 | 0.41 | 1.54 | 0.79–2.24 | 0.15 | ||
| Sex (female/male) | 0.87 | 0.68–1.32 | 0.21 | 0.83 | 0.58–2.43 | 0.42 | ||
| Stage | ||||||||
| II vs. I | 1.85 | 1.82–2.93 | 0.02 | 1.89 | 1.64–3.35 | 0.02 | ||
| III/IV vs. I | 4.24 | 2.55–7.32 | <0.001 | 4.42 | 2.64–8.21 | <0.001 | ||
| (III) GSE37745 (n=105) | ||||||||
| Risk score for 16 metabolism-related genes (high/low risk) | 2.25 | 1.37–3.61 | 0.001 | 1.80 | 1.35–2.47 | 0.001 | ||
| Age (≥65/<65 years) | 1.21 | 0.92–1.65 | 0.112 | 1.02 | 0.76–1.43 | 0.82 | ||
| Sex (female/male) | 0.91 | 0.68–1.32 | 0.21 | 0.86 | 0.53–2.28 | 0.38 | ||
| Stage | ||||||||
| II vs. I | 1.98 | 1.44–2.83 | 0.002 | 1.56 | 1.24–2.47 | <0.001 | ||
| III/IV vs. I | 2.88 | 1.53–4.76 | <0.001 | 2.33 | 1.33–3.91 | <0.001 | ||
The prognostic model consisting of 16 metabolic genes was an independent predictor of survival
Multivariate Cox regression analysis was performed on the prognosis model in the training and validation cohorts to assess whether the predictive power of the model was independent of other clinical factors, such as age, sex and pathological stage. In the multivariate Cox regression analysis, OS was used as the dependent variable, and other clinical factors were used as the covariates. The results showed that after adjusting for other clinical factors, the OS of patients with LUAD were significantly correlated with the prognostic model in the training cohort and validation cohort (training cohort: HR =2.44, 95% CI: 1.58–3.74, P<0.05; validation cohort: HR =2.15, 95% CI: 1.52–2.70, P<0.05) (Table 4, Figure 10A,B).

After assessing the prediction accuracy and independence of the model, we focused on whether a tool combining this model with commonly used clinical factors could benefit patients with LUAD in clinical practice. Therefore, we performed a DCA on the prognostic model to assess the net benefits that the patients might obtain, as shown in Figure 11. The results showed that the combination of the model with age, sex and staging can better predict survival of patients with LUAD in the training cohort and the validation cohort.
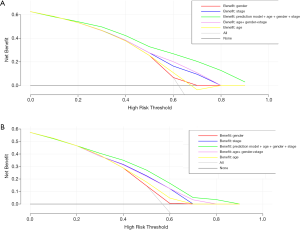
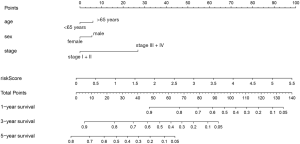
The establishment of the nomograph
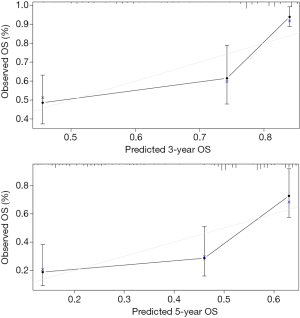
To develop a clinically applicable method to predict the OS of an individual patient, we plotted a nomograph using the prognostic model. The nomograph was plotted after multivariate analysis of the patients with LUAD, as shown in Figure 12. The calibration curves for the 3- and 5-year OS rates suggest that the model has excellent predictive ability for the prognosis of patients with LUAD (3-year survival, 0.701, 5-year survival, 0.685) (Figure 13).
Validation of the prognostic model in the independent LUAD cohorts
We next assessed the predictive power of the prognostic model in the independent LUAD cohorts from the GSE37745 database. In GSE37745, patients were separated into low and high-risk groups based on the calculated risk score, and the OS of the two group was compared. Patients at low risk survived significantly longer than those in high risk (P=0.036, and maximum AUC =0.672). Moreover, the prognostic model consisting of 16 metabolic genes was also an independent predictor of survival in GSE37745 (Table 4, Figure 14A,B).
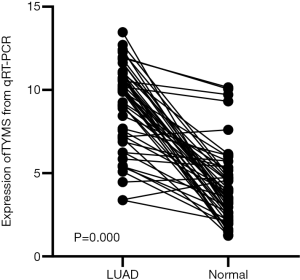
Up-regulation of TYMS mRNA by RT-qPCR
Mapping of PPI networks allowed the construction of a landscape showing how the proteins produced by the genes in our prognostic model. By mapping the whole network, we identified TYMS as the top one hub gene (Figure 15). We analyzed the differential expression of TYMS in 50 LUAD and para-carcinomatous tissue sample pairs through RT-qPCR (Table 5). Expression of TYMS was significantly up-regulated in LUAD tissue of all 50 pairs (9.121±2.543, t=11.079, P<0.001) (Figure 16). Quite notably, expression of TYMS in advanced LUAD was higher than in early stage LUAD (t=−3.584, P=0.001).
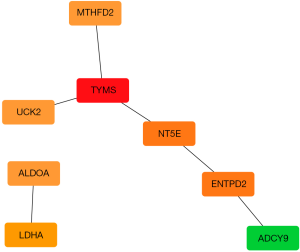
Table 5
| Clinicopathological features | N | TYMS expression (mean ± SD) | T | P |
|---|---|---|---|---|
| Tissues | 11.079 | <0.001 | ||
| Non-cancerous | 50 | 4.11±2.255 | ||
| LUAD | 50 | 9.121±2.543 | ||
| Size | 1.866 | 0.068 | ||
| ≤3 cm | 16 | 8.681±2.689 | ||
| >3 cm | 34 | 10.075±1.948 | ||
| TNM | −3.584 | 0.001 | ||
| I–II | 24 | 7.917±2.606 | ||
| III–IV | 26 | 10.232±1.937 | ||
| Sex | −0.503 | 0.617 | ||
| Male | 26 | 8.946±2.577 | ||
| Female | 24 | 9.311±2.546 | ||
| Age | −1.514 | 0.137 | ||
| ≤65 years | 21 | 8.489±2.864 | ||
| >65 years | 29 | 9.579±2.223 |
LUAD, lung adenocarcinoma; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
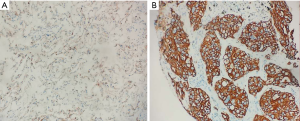
Experimental evidence of TYMS at the protein level by IHC
The staining status of the TYMS antibody was evaluated by an IHC assay and the images of IHC are presented in Figure 17. In 50 LUAD tissues, the number of positive TYMS tissues (68%) was higher than the negative tissues (32%). In the 50 non-cancerous tissues, the number of negative TYMS tissues (70%) was higher than the number of positive tissues (30%). A significant difference in TYMS expression was found between LUAD and non-cancerous lung tissues through Pearson chi-square (χ2=14.44, P=0.0001).
Discussion
Unlimited rapid proliferation is one of the most important biological characteristics of lung cancer cells. To meet the needs of rapid proliferation, tumor cells exhibit metabolic characteristics that are different from those of normal cells: (I) under both oxygen-rich or hypoxic conditions, glycolysis is used as the main energy production mode; (II) a large number of lipids are synthesized to meet the needs of the formation of organelle biofilms in tumor cells and their own special biological functions; and (III) a large number of protein substances are produced to maintain the structure and function of the cells (20,21). In recent years, increasing evidence has indicated that the abnormal expression of metabolic genes is involved in the development and progression of lung cancer. Guo et al. (22) performed high-throughput sequencing of blood samples from patients with LUAD and detected mutations in a large number of metabolic genes. These genes included metabolite transporter genes (CD98 and MCTs), key genes of the tricarboxylic acid cycle (SDH, IDH, and FH), key genes in glucose metabolism (GLUT1 and GPI) and nucleotide anabolism genes (TYMS and RRM2B). In addition, Guo et al. also found that mutations of these metabolic genes were associated with the progression of NSCLC. Pérez-Ramírez et al. (23) also showed that NSCLC patients with MDM2 mutations had a shorter OS. To date, hundreds of proteins have been considered to be involved in metabolic processes. Given the importance of metabolic genes in lung cancer, we speculate that metabolic genes may have certain importance in patient prognosis and can be used as ideal markers for prognoses.
In this study, we analyzed the mRNA expression of 455 metabolic genes in the TCGA dataset. Among them, 59 genes were associated with the survival of patients with LUAD. We established a prognostic model consisting of 16 genes using LASSO regression. The risk scores for each patient were calculated by integrating the mRNA expression levels and regression coefficients of selected genes. The prognostic model divided the patients with LUAD into a high-risk group and a low-risk group, and the OS for the high-risk and low-risk groups was significantly different. More importantly, the prognostic model composed of the above 16 genes was also validated in a validation cohort with 189 patients. Over the past decade, many researchers have developed prognostic models related to clinical disease outcomes. These models, also known as classifiers, are often used to predict the prognosis of patients with various diseases (24-26). Some models are even better than TNM staging in predicting prognosis. Currently, prognostic models based on metabolic genes have been reported in liver cancer and breast cancer. For example, Liu et al. (27) recently reported a predictive prognostic model consisting of four metabolic genes (ACAT1, GOT2, PTDSS2 and UCK2) to predict OS in patients with hepatocellular carcinoma (HCC). This prognostic model successfully separated patients with significantly increased early recurrence risk from patients with low risk. Comparing the model established in this study with the model developed by Liu, we found that the UCK2 gene appeared in both models, indicating that the UCK2 gene plays a role in both HCC and LUAD. UCK2 is a pyrimidine nucleotide kinase capable of phosphorylating uridine and cytosine to uridine monophosphate and cytidine phosphate monophosphate. UCK2 is the rate-limiting enzyme in the salvage pathway of pyrimidine nucleotides. When a tumor develops, various physiological functions of the body become disordered, and the normal de novo synthesis of nucleotides may be affected to some extent. Therefore, the amount of generated nucleotides cannot satisfy the rapid proliferation and strong metabolism of tumor cells. The existence of salvage pathways meets the needs of tumor proliferation. Therefore, the expression of UCK2 was elevated in both HCC and LUAD. Huang et al. (28) also showed that UCK2, as an oncogene, promoted the proliferation and invasion of HCC cells. Knocking out UCK2 inhibited the proliferation and colony formation ability of tumor cells. Wu et al. (29) also showed that UCK2 promoted the proliferation of LUAD cells and might be involved in the MTOR1 signaling pathway or interacts with the MYC or E2F genes.
To evaluate the prediction accuracy of the prognostic model, we performed time-dependent ROC curve analysis and calculated the AUC values at different cut-off times. The AUC value of the 3-year survival rate was predicted to be 0.732, and the AUC value of the 5-year survival rate was 0.69 in the validation cohort. The above results are consistent with those in the training cohort, demonstrating excellent prediction accuracy of the model. However, AUC only focuses on the prediction accuracy of the model and does not suggest whether the model is worthy of clinic application. DCA is a statistical method that contains results and thus can provide information for whether the model can be used in the clinical practice (30). We applied DCA to evaluate our prognostic model and found that the combination of the prognostic model with clinical parameters such as age, sex and staging can benefit patients with LUAD the most.
Using the prognostic model, we found that the expression of ALDOA, LDHA, PTGES, and MTHFD2 was relatively high in high-risk patients and that the expression of the above genes was low in low-risk patients. ALDOA is a glycolytic enzyme and an essential enzyme for ATP synthesis that can promote the invasiveness of tumor cells by regulating the wnt signaling pathway (31). LDHA is associated with the progression of breast cancer, lung cancer, and glioma (32,33). Wang et al. (34) found that PTGES, a key enzyme that synthesizes PGE2 in the arachidonic acid pathway, is highly downregulated in NSCLC. The abnormal expression of PTGES is critical for the promotion of lung cancer cell migration and metastasis. This is related to the imbalance of protein stability by the deubiquitinating enzyme USP9X. MTHFD2 is a bifunctional enzyme located in mitochondria and is overexpressed in various malignant tumors. Yu et al. (35) showed that MTHFD2 knockout inhibited the proliferation of lung cancer cells by mediating the inhibition of cell cycle-related genes, providing a new basis for targeted therapy of NSCLC.
Bioinformatics studies showed that the 16 genes in the LUAD prognostic model were mainly related to purine metabolism, pyrimidine metabolism, glycolysis and other signaling pathways, indicating that multiple metabolic pathways undergo significant changes during cell malignant transformation. The involved pathways are the tricarboxylic acid cycle, oxidative phosphorylation, amino acid metabolism, fatty acid metabolism and nucleic acid metabolism. Cell malignant transformation is therefore also known as reprogramming of energy metabolism in tumor cells, usually caused by gene mutations (36,37). Interestingly, this study found that metabolic genes also play roles in tumor drug metabolism pathways, suggesting that metabolic genes may be associated with antitumor drug resistance. This study also found that a large number of metabolic genes are involved in the glycolysis pathway. Glucose metabolism is the main energy source of cells. Warburg confirmed that due to the rapid growth of tumor cells, glycolysis can still be used for glycolysis in the presence of adequate oxygen. Complex factors such as the microenvironment and gene mutations can cause changes in glucose metabolism in tumor cells. First, it can cause the upregulation of glycolysis-associated enzymes and GLUT; on the other hand, the tricarboxylic acid cycle is inhibited, and the aerobic oxidation of mitochondria is weakened, resulting in increased glycolytic activity (38). Zhang et al. (4) suggested that glycolytic genes were associated with the poor prognosis of patients with LUAD, which is consistent with the findings of our study.
It is worth noting that purine nucleotide metabolic pathways are the most closely related to metabolic genes, indicating the imbalance of purine nucleotide metabolism in the process of lung cancer occurrence. Rampazzo et al. (39) showed that the antitumor mechanisms of gemcitabine involve competitive inhibitory effects on metabolic enzymes that interfere or inhibit the synthesis of purine nucleotides. A large amount of CN-II enzymes on the surface of tumor cells can cause the dephosphorylation of nucleoside analogs and thus drug resistance. Yuan (40) suggested that serum uric acid plays an antioxidant role but that increased serum uric acid is correlated with an increased risk of lung cancer and mortality. Uric acid has complex biological functions in lung cancer. The prognostic model developed in this study also included the ADCY9 gene, which is involved in purine metabolism. ADCY9 is a member of the membrane-bound adenylate cyclase family and plays an important regulatory role in cell proliferation and differentiation. Tan et al. (41) showed that ADCY9 gene mutations reduced the amount of mitochondrial DNA, suggesting that ADCY9 plays a role in providing dNTPs for mitochondrial DNA synthesis, thus affecting the proliferation and metastasis of lung cancer. However, the specific mechanisms and pathways that affect the proliferation and metastasis of LUAD by ADCY9 still require further basic research.
Finally, we developed a nomograph to predict the OS of an individual patient. A nomograph is a stable and reliable tool for quantitative analysis by combining and depicting risk factors. It has been widely used to assess the prognosis of various diseases, including lung cancer (42). A nomograph generates a statistical prediction model that is presented as a graph to assign values to specific clinical factors (e.g., age, sex and staging). By summarizing all the points, a nomograph can provide an individual patient with a digital display of prognoses, such as OS, time to relapse, and interval of acute exacerbation. In addition to traditional clinical features (TNM staging, tumor size and target gene mutations), risk scores calculated using prognostic models can also be included in predictive nomographs to better predict prognosis. Li et al. (19) reported a nomograph for predicting the 1-, 3-, and 5-year OS of patients with LUAD. The risk score was calculated based on gene expression in the model. The results suggested that the combination of risk scores and traditional prognostic factors can more accurately predict prognosis, which is consistent with the findings of our study. The calibration curve established in this study showed that the nomograph combining the prognostic model consisting of metabolic genes and conventional prognostic factors could accurately predict the 3- and 5-year survival of patients with LUAD. When the same prognostic model was used to calculate risk scores in GSE37745, we found that this model also could provide with high prediction ability.
In order to further verify the accuracy of the model, we choose TYMS gene as a representative metabolic gene using PPI network. Then, we verify the expression of TYMS in LUAD tissues by RT-qPCR and immunohistochemical staining. Expression of TYMS was significantly up-regulated in LUAD tissue of all 50 pairs. Moreover, expression of TYMS in advanced LUAD was higher than in early stage LUAD. A significant difference in TYMS expression was also found between LUAD and non-cancerous lung tissues using immunohistochemical staining. TYMS is the enzyme which catalyze the reaction that provides the sole de novo intracellular source of thymidylate, which is important for Folic acid metabolism and DNA synthesis. Elevated TYMS expression or activity may play a part in the proliferation and metastasis of lung cancer.
In summary, based on the TCGA dataset, we constructed a prognostic model consisting of 16 metabolic genes. This model represents an independent prognostic factor. The nomograph combining the prognostic model and conventional prognostic factors could accurately predict the 3- and 5-year survival of induvial patients with LUAD. This study may provide guidance for the individualized treatment of patients with lung cancer.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1571). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tiansheng G, Junming H, Xiaoyun W, et al. lncRNA Metastasis-Associated LUAD Transcript 1 Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer Cells via Down-Regulating miR-202 Expression. Cell J 2020;22:375-85. [PubMed]
- Rochigneux P, Garon EB. Are lung adenocarcinoma mutations shaping the immune microenvironment? Transl Cancer Res 2018;7:S740-2. [Crossref] [PubMed]
- Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol 2012;13:528-38. [Crossref] [PubMed]
- Zhang L, Zhang Z, Yu Z. Identification of a novel glycolysis-related gene signature for predicting metastasis and survival in patients with LUAD. J Transl Med 2019;17:423. [Crossref] [PubMed]
- Carter BW, Lichtenberger JP 3rd, Benveniste MK, et al. Revisions to the TNM Staging of Lung Cancer: Rationale, Significance, and Clinical Application. Radiographics 2018;38:374-91. [Crossref] [PubMed]
- Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17:351-9. [Crossref] [PubMed]
- Weiss RH. Metabolomics and Metabolic Reprogramming in Kidney Cancer. Semin Nephrol 2018;38:175-82. [Crossref] [PubMed]
- Abbaszadeh Z, Cesmeli S, Biray Avci C. Crucial players in glycolysis: Cancer progress. Gene 2020;726:144158. [Crossref] [PubMed]
- Brown TP, Ganapathy V. Lactate/GPR81 signaling and proton motive force in cancer: Role in angiogenesis, immune escape, nutrition, and Warburg phenomenon. Pharmacol Ther 2020;206:107451. [Crossref] [PubMed]
- Liu Y, Bai F, Liu N, et al. The Warburg effect: A new insight into atrial fibrillation. Clin Chim Acta 2019;499:4-12. [Crossref] [PubMed]
- Avnet S, Baldini N, Brisson L, et al. Annual Meeting of the International Society of Cancer Metabolism (ISCaM): Metabolic Adaptations and Targets in Cancer. Front Oncol 2019;9:1332. [Crossref] [PubMed]
- Lebelo MT, Joubert AM, Visagie MH. Warburg effect and its role in tumourigenesis. Arch Pharm Res 2019;42:833-47. [Crossref] [PubMed]
- Cassim S, Pouyssegur J. Tumor Microenvironment: A Metabolic Player that Shapes the Immune Response. Int J Mol Sci 2019; [Crossref] [PubMed]
- Yun J, Rago C, Cheong I, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009;325:1555-9. [Crossref] [PubMed]
- Hu S, Balakrishnan A, Bok RA, et al. 3C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 2011;14:131-42. [Crossref] [PubMed]
- Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19:17-30. [Crossref] [PubMed]
- Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res 2013;19:194-204. [Crossref] [PubMed]
- Zeng L, Wang W, Chen Y, et al. A five-long non-coding RNA signature with the ability to predict overall survival of patients with LUAD. Exp Ther Med 2019;18:4852-64. [PubMed]
- Li Y, Ge D, Gu J, et al. A large cohort study identifying a novel prognosis prediction model for LUAD through machine learning strategies. BMC Cancer 2019;19:886. [Crossref] [PubMed]
- Takada K, Toyokawa G, Okamoto T, et al. Metabolic characteristics of programmed cell death-ligand 1-expressing lung cancer on (18) F-fluorodeoxyglucose positron emission tomography/computed tomography. Cancer Med 2017;6:2552-61. [Crossref] [PubMed]
- Lim SL, Jia Z, Lu Y, et al. Metabolic signatures of four major histological types of lung cancer cells. Metabolomics 2018;14:118. [Crossref] [PubMed]
- Guo X, Li D, Wu Y, et al. Genetic variants in genes of tricarboxylic acid cycle key enzymes are associated with prognosis of patients with non-small cell lung cancer. Lung Cancer 2015;87:162-8. [Crossref] [PubMed]
- Pérez-Ramírez C, Cañadas-Garre M, Alnatsha A, et al. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenomics J 2019;19:164-77. [Crossref] [PubMed]
- Tan W, Xie X, Huang Z, et al. Construction of an immune-related genes nomogram for the preoperative prediction of axillary lymph node metastasis in triple-negative breast cancer. Artif Cells Nanomed Biotechnol 2020;48:288-97. [Crossref] [PubMed]
- Wang J, Wang Y, Kong F, et al. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J Cell Physiol 2020;235:3056-68. [Crossref] [PubMed]
- Lv X, Zhao Y, Zhang L, et al. Development of a novel gene signature in patients without Helicobacter pylori infection gastric cancer. J Cell Biochem 2020;121:1842-54. [Crossref] [PubMed]
- Liu GM, Xie WX, Zhang CY, et al. Identification of a four-gene metabolic signature predicting overall survival for hepatocellular carcinoma. J Cell Physiol 2020;235:1624-36. [Crossref] [PubMed]
- Huang S, Li J, Tam NL, et al. Uridine-cytidine kinase 2 upregulation predicts poor prognosis of hepatocellular carcinoma and is associated with cancer aggressiveness. Mol Carcinog 2019;58:603-15. [Crossref] [PubMed]
- Wu Y, Jamal M, Xie T, et al. Uridine-cytidine kinase 2 (UCK2): A potential diagnostic and prognostic biomarker for lung cancer. Cancer Sci 2019;110:2734-47. [Crossref] [PubMed]
- Liu GM, Zeng HD, Zhang CY, et al. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int 2019;19:138. [Crossref] [PubMed]
- Chang YC, Chiou J, Yang YF, et al. Therapeutic Targeting of Aldolase A Interactions Inhibits Lung Cancer Metastasis and Prolongs Survival. Cancer Res 2019;79:4754-66. [PubMed]
- Jin L, Chun J, Pan C, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 2017;36:3797-806. [Crossref] [PubMed]
- Deme D, Telekes A. Prognostic importance of lactate dehydrogenase (LDH) in oncology. Orv Hetil 2017;158:1977-88. [Crossref] [PubMed]
- Wang T, Jing B, Sun B, et al. Stabilization of PTGES by deubiquitinase USP9X promotes metastatic features of lung cancer via PGE2 signaling. Am J Cancer Res 2019;9:1145-60. [PubMed]
- Yu C, Yang L, Cai M, et al. Down-regulation of MTHFD2 inhibits NSCLC progression by suppressing cycle-related genes. J Cell Mol Med 2020;24:1568-77. [Crossref] [PubMed]
- Biswas SK. Metabolic Reprogramming of Immune Cells in Cancer Progression. Immunity 2015;43:435-49. [Crossref] [PubMed]
- López-Sánchez LM, Aranda E, Rodríguez-Ariza A. Nitric oxide and tumor metabolic reprogramming. Biochem Pharmacol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 2013;12:152. [Crossref] [PubMed]
- Rampazzo C, Tozzi MG, Dumontet C, et al. The druggability of intracellular nucleotide-degrading enzymes. Cancer Chemother Pharmacol 2016;77:883-93. [Crossref] [PubMed]
- Yuan X. Research progress on the correlation of purine nucleotide metabolism and malignant tumors. Oncology Progress 2019;17:2498-501.
- Tan X, Banerjee P, Liu X, et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest 2018;128:1267-82. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]