
Biological effects of ubiquitin-specific peptidase 22 on thyroid papillary cancer cells and its mechanism of action
Introduction
In recent years, advances in medical science and technology have led to an annual increase in the incidence and detection rates of thyroid papillary cancer (TPC), with younger adults being particularly affected. Therefore, more extensive study into TPC to gain a better understanding of the mechanism underlying its occurrence and development is urgently needed to inform better prevention and treatment strategies.
Ubiquitin-specific peptidase 22 (USP22) is a member of the deubiquitinating enzyme (DUB) family. Exerting a wide range of biological functions, USP22 mainly depends on the deubiquitination of substrate target molecules and plays a key role in cell cycle regulation. The USP22 gene is located at chromosome 17p11.2 and comprises 14 exons. Its DNA contains a 1,578 bp open reading frame, encoding a polypeptide containing 525 amino acids. It has a relative molecular weight (Mr ×103) of about 60 kDa. High expressions of USP22 have recently been found in many tumors, such as liver cancer, glioma, and lung cancer (1-4). These findings confirmed the crucial role USP22 serves in the invasion and metastasis of cancer cells as a proto-oncogene. Glinsky et al. (5). identified and summarized a set of death-from-cancer signatures including 11 genes, which are expressed continually as stem cells in primary tumors and metastatic tissues and are closely related to tumor occurrence and development, distant metastasis, drug resistance, and recurrence after surgery. There are two tagging genes: USP22 and multicomb Bmi-1.
The role performed by USP22 in TPC has yet to be established. In this study, USP22 was detected in TPC and normal thyroid epithelial cell lines, and its biological function and molecular mechanism were analyzed to provide new evidence for the prevention, diagnosis, treatment, and prognosis of TPC.
Methods
Materials
Antibodies and consumables
The following primary antibodies were purchased: β-actin monoclonal antibody (60008-1-Ig, Proteintech in America), USP22 polyclonal antibody (LS-C99567, LifeSpace in Australia); Bmi-1 polyclonal antibody (ab126783, Abcam in England); and cyclin D2 polyclonal antibody (TA323121, Origene Company in America).
The secondary antibodies were sheep anti-rabbit IgG antibodies and sheep anti-mouse IgG antibodies, purchased from the ThermoFisher Scientific (America). Some reagents and consumables were obtained from Beijing Zhongshan Jinqiao Biotechnology Co. Ltd. (China).
Cell lines
TPC-1 (BNCC337912) and human normal thyroid cell line HT-ori3 (BNCC 338687) were purchased from Beina Chuanglian Biotechnology Co. Ltd. (China).
Primer sequence
| Primer F (5'-3') | Primer R (3'-5') | |
|---|---|---|
| siRNA-1 | GGAGAAAGAUCACCUCGAATT | TTCCUCUUUCUAGUGGAGCUU |
| siRNA-2 | GCAUCAUAGACCAGAUCUUTT | TTCGUAGUAUCUGGUCUAGAA |
| Negative control | UUCUCCGAAGGUGUCACGUTT | TTAAGAGGCUUGCACAGUGCA |
| Primer F (5'-3') | Primer R (3'-5') | |
|---|---|---|
| USP22 | CACTTCTGCGGGACT-3 | TACGGGATGTGAGGG |
| GAPDH | AGCAAGAGCACAAGAGGAAG | GGTTGAGCACAGGGTACTTT |
Study methods
Cell culture, resuscitation, and passage
Cell transfection
Cells of TPC-1 and HT-ori3 underwent a series of treatments. First, 6 µL of siRNA-NC, siRNA-1, and siRNA-2 was added to the first row of Eppendorf (EP) tubes respectively, and 3 µL of Lipofectamin 3000 transfection reagent was added to the other row of EP tubes. Next, the liquid of 2 rows EP tubes were mixed respectively, and incubated for 15 min. The mixture was added to both the control cells and the experimental cells, and then shaken and cultured in an incubator. After 48 h, experiments were performed on transient cell lines.
Cell function tests
(I) 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay for cell viability
The digested cells were cultured in 5 96-well plates (1,000 cells/well) containing 100 µL of Dulbecco’s Modified Eagle Medium (DMEM)/Roswell Park Memorial Institute (RPMI) 1640 Medium. Each group had six repetitive wells. The culture medium was discarded after 24 h. Next, 100 µL of MTT solution (MTT:medium =1:9) was added to each well, and the cells was incubated for 2 h. The MTT solution was then discarded. After that, 100 µLdimethyl sulfoxide (DMSO) was added to each well, and the plates were shaken for 15 min in the dark. Absorbance (OD570) was measured for five consecutive days. Finally, the cell viability in the control group and the USP22-siRNA1 group was compared.
(II) Migration and invasion assay
TPC-1 cells of the USP22-siRNA1 group and control group were starved for 12 h. For the invasion assay, matrigel matrix was laid 4 h on the Transwell cabin in advance, and 60 µL of mixture was added to the upper chamber, and then put at 37 °C in an incubator. The migration experiment was conducted without matrigel matrix. Starved cells (1×104) were resuspended in the upper chamber, and cultured in an incubator. If adherent cells were observed in the lower chamber, the culture was terminated. The small chamber was taken out, observing adherent cells and fixed in 600 µL 90% ethanol. After 45 min, the culture medium was discarded, dried for 10 min, and stained with 600 µL 0.1% crystal violet for 10 min. The cells were washed three times with PBS and observed and photographed under a microscope.
(III) Colony formation assay
After 48 h of transfection, cells were digested and seeded into 6-well plates with 1,000 cells/well. Three wells were repeated in every group. The solution was changed every 2–3 days until clones were clearly visible under the microscope. After 10 days, the cells were harvested, fixed, and stained with 0.1% crystal violet. The supernatant was discarded, and the cells were washed with water, and photographed.
Western blot
(i) Cells of six-well plate were added with lysis buffer[mixed 1% phenylmethylsulfonyl fluoride (PMSF), a protease inhibitor]. Scraped cells from every well were placed on ice for 30 min. After centrifuging, to which 5 × loading buffer with 2-mercaptoethanol was added. Then, the tubes were heated at 100 °C for 10 min. (ii) Water was added to the gel-prepared plates and left for 5 min, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis(SDS-PAGE) was prepared. (iii) Ten percent of the separation gels were poured into the interspace of the gel-prepared plates, left for 45 min until coagulation occurred. (iv) Five percent concentrated gel was added. The comb was inserted, and stood for 1–2 h until coagulation occurred. Next, the comb was pulled out, and 3 µL of protein marker was added. Electrophoresis initially commenced with a low voltage of 60 V. When the marker strip was separated and appeared red, the voltage was increased to 120 V. (v) After electrophoresis, wet membrane transfer was conducted for 90 min. The polyvinylidene fluoride (PVDF) membrane was rinsed with water, and then soaked in Ponceau S. The Ponceau S was collected, the cells were rinsed, and the target strip was cut and placed in 5% skim milk powder [20 mL phosphate buffer solution-tween (PBST) + 1 g milk] and sealed for 1.5 h on the shaker. The target strip was washed and incubated with the primary antibody at 4 °C overnight. Then, the target strip was washed. After incubation with second antibody for 1 h, the target strip was briefly washed. Finally, developer was added to the membrane and it was photographed for later use.
Extraction of RNA and qPCR
RNA extraction
RNA was extracted using Trizol (Thermo Fisher Scientific, in America) first. Chloroform was added to each EP tube. The samples were kept at room temperature for 15 min, before centrifugation at 4 °C at 11,000 g for 15 min. The mixed solution was divided into three layers. The upper layer of the solution was transferred to a 1.5-mL EP tube without RNA enzymes and added to isopropanol with 80% of the total volume of the upper layer. The mixture was kept for 5 min, and then centrifuged at 4 °C at 11,000 g for 15 min. The supernatant was discarded. Next, salt was removed by adding 1 mL of 70% pre-cooled alcohol, the mixture was centrifuged at 8,000 g at 4 °C for 10 min, and the supernatant was discarded and dried. When the sediment became transparent, 60 µL diethyl pyrocarbonate (DEPC) water was added, the samples were loaded separately and stored at −80 °C.
RT-PCR 42 °C 15 min → 85 °C 5 s → 4 °C save
q-PCR two-step method: 94 °C 30 s → 94 °C 15 s→ 60 °C 30 s → 95 °C 15 s → 60 °C 1 min → 95 °C 15 s, 40 cycles.
Statistical analysis
Statistical analyses were performed using SPSS version 17.0 software (International business machines corporation in America). All data were expressed as mean ± standard deviation), and the data between groups was compared using the t-test. P<0.05 was considered to be statistically significant.
Results
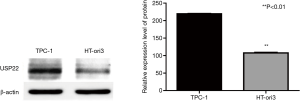
USP22 protein expression in TPC-1 and HT-ori 3 cells
Expression of USP22 protein in TPC-1 cells was higher than that in HT-ori 3 normal thyroid epithelia l cells (P<0.01, Figure 1). The USP22 protein band of Weserblot was darker in TPC-1 cells. The USP22 protein level of TPC-1 cells was about twice of HT-ori 3 cells (Figure 1).
The effects of USP22 gene on the biological behavior of TPC-1 cells
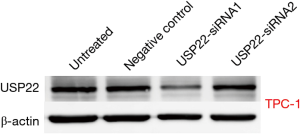
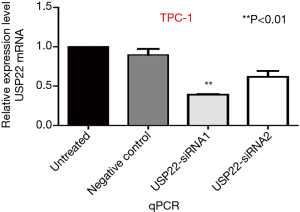
USP22 gene silencing
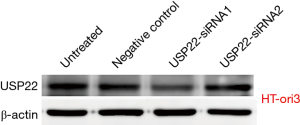
TPC-1 cells were transfected with two types of specific siRNA targeting USP22. The expression levels of USP22 protein and mRNA in the USP22-siRNA1 group were significantly lower than those in the negative control and untreated groups (P<0.01, Figures 2 and 3). Therefore, USP22-siRNA1 was selected for the subsequent experiments.
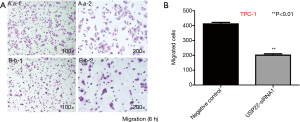
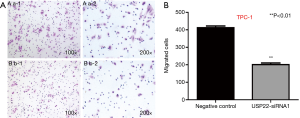
Transwell cell migration assay
Cell migration assay showed that 6 h after USP22 gene silencing, the number of TPC-1 cells was decreased in the USP22-siRNA1 group compared with the negative control group [402/422 (/lower power field, LPF, 10×10) in the control group vs. 192/210 (/LPF) in the USP22-siRNA1 group]. There were significant differences between the two groups (P<0.01, Figure 4). Morphological observation showed that the cells in the USP22-siRNA1 group had become smaller and the cytoplasm was more concentrated (Figure 4).
Transwell cell invasion assay
Transwell assay showed that 24 hours after gene silencing, the number of invasive cells was decreased in the USP22-siRNA1 group compared with the negative control group [128/141 (/LPF) in the TPC-1 control group vs. 73/80 (/LPF) in the USP22-siRNA1 group]. There were significant differences between the two groups (P<0.05, Figure 5). The cells were also reduced in size in the USP22-siRNA1 group.
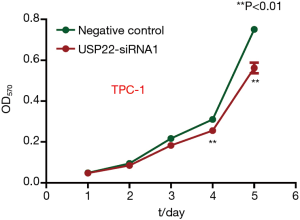
MTT assay
To compare cell viability in the control group and USP22-siRNA1 group, an MTT assay was performed for five consecutive days. The difference in OD570 value between the two groups increased significantly on days 4 and 5. The OD570 values are shown in Figure 6. There was a significant difference between the two groups (P<0.01, Figure 6).
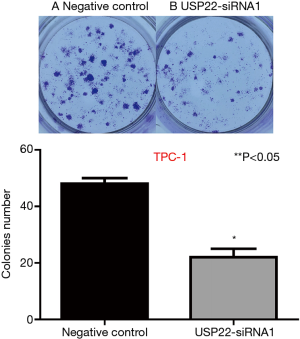
Clone formation assay
After 10 days of gene silencing, the number of clones formed by the TPC-1 cells was 46/50 in the control group and 19/25 in the USP22-SiRNA1 group. There was a significant difference between the two groups (P<0.05, Figure 7).
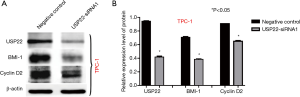
USP22, CyclinD2 and Bmi-1 protein levels
After gene silencing, the protein levels of cyclin D2, Bmi-1, and USP22 in the TPC-1 cell lines were measured by Western blotting. Compared with the control group, the protein levels of USP22, cyclin D2, and Bmi-1 were significantly decreased (P<0.05, Figure 8A). Meanwhile, the decreased protein levels of USP22, cyclin D2, and Bmi-1 were positively related (P<0.05, Figure 8B).
The effects of gene silencing on normal thyroid cells
USP22 gene silencing
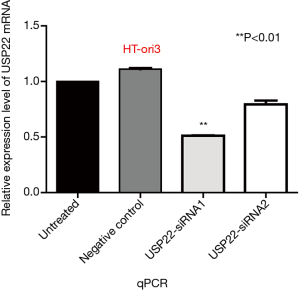
HT-ori3 normal thyroid epithelial cells were transfected with two kinds of specific siRNA targeting USP22. The expression levels of USP22 protein and mRNA in the USP22-siRNA1 group were significantly lower than those in the negative control and untreated groups (P<0.05, Figures 9 and 10). Therefore, USP22-siRNA1 was selected for the subsequent experiments.
MTT assay
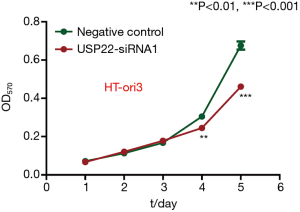
To compare cell viability between the control group and experimental group, an MTT assay was performed for five consecutive days. The difference in OD570 between the two groups increased significantly on days 4 and 5 (Figure 11), There was a significant difference between the two groups (P<0.01).
Clone formation assay
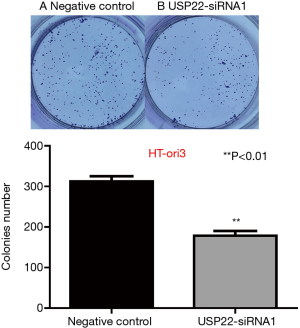
After 10 days of gene silencing, the number of small clones formed by the HT-ori3 cells was 325/300 in the negative control group compared with 190/167 in the experimental USP22-siRNA1 group. There was a significant difference between the two groups (P<0.01, Figure 12).
Discussion
The proteins encoded by USP22 are expressed in the nucleus. The histidine (His) and cysteine (Cys) at the carboxyl end of USP22 are responsible for ubiquitin hydrolase activity and can remove ubiquitin from macromolecular proteins. The amino end has a zinc finger structure and forms the structural basis for mediating the interaction between the molecule itself and the target protein of the substrate. Moreover, USP22 needs to interact with other auxiliary molecules to form a complex and exert its biological effects (6), and this biological function is dependent on the integrity of the TFTC/STAGA complex. Yi et al. (7) found that USP22 expression was low in normal cells and high in thyroid cancer cell lines, which was related to cell proliferation. In vivo experiments showed by downregulating USP22, the tumorigenic activity of thyroid cancer cells in nude mice was inhibited and the expression of cell-cycle-related molecules was decreased. In the present study, we found that expression of USP22 in TPC-1 cancer cells was significantly higher than that in normal cells, suggesting that USP22 is closely related to tumorigenesis, which is consistent with the findings of Yi et al. (7).
In the present study, after silencing of the USP22 gene in TPC-1 cells, after 6 h the migration rate was lower and the cells had become smaller in size compared with the control group. The number of invasive cells at 24 h was significantly lower in the experimental group than in the control group. The cell viability test showed no significant difference between the experimental and control groups on days 1 and 2, but on days 4 and 5, the difference increased. The number of colony-forming cells was also significantly decreased in the experimental group after 10 days compared with the control group. In the experimental group, the levels of USP22 gene and protein expression were significantly decreased (both lower than in the control group), which indicates that the USP22 gene plays a key role in the growth, proliferation, invasion, and migration of TPC cells.
USP22 plays an important role in TPC and in the growth of normal thyroid cells. In the present study, after silencing of the USP22 gene in normal HT-ori3 cells, the levels of USP22 gene and protein expression were significantly decreased (both significantly lower than in the negative control group). The cell viability test showed that no significant difference existed between the HT-ori3 cells and the negative control group on days 1 and 2. However, the difference both increased on days 4 and 5, which was significantly lower in the experimental group than in the negative control group (P<0.01 on day 4 and P<0.001 on day 5). Meanwhile, the number of cloned cells was significantly decreased in the experimental group after 10 days compared with the negative control group (P<0.01). These findings suggest that the USP22 gene and protein also play an indispensable role in the growth and development of normal thyroid cells.
Previous studies have found that the decrease in USP22 expression in cancer cells can lead to cell cycle arrest, retention of cells in the G1 phase, and inhibition of cell proliferation (4,8-10), which may be related to the cyclin D2, Bmi-1, and p53 signaling pathways (8). Cyclin D2 is the key cyclin from stages G1 to S; therefore, the effect of USP22 on cell proliferation and cell cycle may be accomplished through cyclin D2. Many experiments have confirmed that Bmi-1 and USP22 have overlapping functions (10). The expression levels of USP22 and Bmi-1 in colorectal cancer tissues are significantly higher than those in the adjacent normal tissues, and there is a significant positive correlation between them. The co-activation of USP22 and Bmi-1 may promote the progression of colorectal cancer and be a predictor of poor prognosis (11). In this study, we observed that after USP22 gene silencing, the protein levels of USP22 and its corresponding proteins Bmi-1 and cyclin D2 were significantly lower than those of the control group. The decrease of USP22 was positively correlated with the decrease of Bmi-1 and cyclin D2, which suggests that USP22 exerts its effect in TPC via the Bmi-1 and cyclin D2 pathways.
In recent years, different ideas regarding the role of USP22 have been put forward by various tumor and organizational research studies. USP22 deletion in endothelial cells and pericytes of the mouse placenta has been shown to hinder some signaling cascades with primary effects on cell survival, differentiation, and the ability to form vessels, including TGFβ and several receptor tyrosine kinase (RTK) pathways (12). USP22 deficiency was also observed to lead to myeloid leukemia upon oncogenic Kras activation through a PU.1-dependent mechanism (13). Huang et al. (14) identified USP22 as a novel deubiquitinase of CD274 (PD-L1), which directly interacted with the C terminus of PD-L1 and improved therapeutic efficacy of PD-L1-targeted immunotherapy and CDDP-based chemotherapy in mice. Moreover, genetic depletion of USP22 increased tumor immunogenicity and tumor-infiltrating lymphocytes. Meanwhile, Zhao et al. (15) showed that USP22 silencing restrained cell migration and invasion by inhibiting epithelial-mesenchymal transition in in vitro assays; knockdown of USP22 promoted mitochondrion-mediated and caspase-dependent apoptosis via the upregulation of Bax and Bid and the promotion of caspase-3 activation. Meanwhile, downregulation of USP22 in anaplastic thyroid carcinoma cells may impede lung metastasis in vivo.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2102
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2102). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ding F, Bao C, Tian Y, et al. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int J Mol Sci 2014;16:307-20. [Crossref] [PubMed]
- Machida YJ. A mechanism for the tissue specificity in BAP1 cancer syndrome. Transl Cancer Res 2019;8:S621-4. [Crossref]
- Liang J, Zhang XL, Li S, Xie S, Wang WF, Yu RT. Ubiquitin-specific protease 22 promotes the proliferation, migration and invasion of glioma cells. Cancer Biomark 2018;23:381-9. [Crossref] [PubMed]
- Zhuang YJ, Liao ZW, Yu HW, et al. ShRNA-mediated silencing of the ubiquitin-specific protease 22 gene restrained cell progression and affected the Akt pathway in nasopharyngeal carcinoma. Cancer Biol Ther 2015;16:88-96. [Crossref] [PubMed]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503-21. [Crossref] [PubMed]
- Lin Z, Yang H, Kong Q, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell 2012;46:484-94. [Crossref] [PubMed]
- Yi J. USP22 promotes growth of thyroid carcinoma. Fourth Military Medical University 2014;01:3-59.
- Liu YL, Zheng J, Tang LJ, et al. The deubiquitinating enzyme activity of USP22 is necessary for regulating HeLa cell growth. Gene 2015;572:49-56. [Crossref] [PubMed]
- Kobayashi T, Iwamoto Y, Takashima K, et al. Deubiquitinating enzymes regulate Hes1 stability and neuronal differentiation. FEBS J 2015;282:2411-23. [Crossref] [PubMed]
- Liu YL, Jiang SX, Yang YM, et al. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys 2012;62:229-35. [Crossref] [PubMed]
- Sun ZC, Cui BB, Liu YL, et al. Clinical Significance of the Expression of USP22 and BMI-1 in Colorectal Cancer. Progress in Modern Biomedicine. Xian Dai Sheng Wu Yi Xue Jin Zhan 2014;33:6432-6.
- Koutelou E, Wang L, Schibler AC, et al. USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development 2019;146:dev174037. [Crossref] [PubMed]
- Melo-Cardenas J, Xu Y, Wei J, et al. USP22 deficiency leads to myeloid leukemia upon oncogenic Kras activation through a PU.1-dependent mechanism. Blood 2018;132:423-34. [Crossref] [PubMed]
- Huang X, Zhang Q, Lou Y, et al. USP22 Deubiquitinates CD274 to Suppress Anticancer Immunity. Cancer Immunol Res 2019;7:1580-90. [Crossref] [PubMed]
- Zhao HD, Tang HL, Liu NN, et al. Targeting ubiquitin-specific protease 22 suppresses growth and metastasis of anaplastic thyroid carcinoma. Oncotarget 2016;7:31191-203. [Crossref] [PubMed]


