
ARID1A/BAF250a is significantly overexpressed in primary invasive breast cancer
Introduction
Cancer development and progression may result from genomic as well as epigenetic alterations (1-3). In a variety of human cancers, epigenome is affected by mutations in some genes, which are involved in the regulation of the chromatin structure (4,5). The chromatin structure is established by two major groups of functional units, which interact dynamically with each other. The first group of complexes modifies the association of DNA and histone octamers involving chemical energy derived from ATP hydrolysis. The latter one modulates histone N-terminal tails in a covalent manner (5,6).
ATP-dependent remodelers were first discovered in 1984 in studies of yeast (Saccharomyces cerevisiae) as SWI/SNF (SWItch/Sucrose Non Fermentable) complexes. A few years later that discovery was followed by a detection of corresponding complexes in Drosophila (7).
BAF250a (also known as ARID1A, p270 or SMARCF1) is a key component of the mammalian SWI/SNF family, it cooperates with transcriptional activators and repressors, and in this manner triggers the recruitment of the remodeling complex and its activity (8-10). Dysregulated activation of ATP-dependent chromatin remodelers can be detected in a variety of tumors. The BAF250a subunit is the most frequently affected among all SWI/SNF subunits (9). ARID1A gene mutations have been found in many types of malignancies, including breast cancer (4,10).
Our study was designed to evaluate the expression of BAF250a in 119 females diagnosed with primary breast cancer by immunohistochemical staining (11). The results were correlated with clinicopathological features such as age, breast cancer type, menopause, tumor size and location, and lymph nodes metastasis.
Methods
Patients
Ethics approval (number: KB 132/2018) was obtained by the Bioethics Committee of the Wroclaw Medical University. The study was performed using formalin-fixed and paraffin-embedded (FFPE) blocks of 119 primary breast cancer tissues and 92 healthy controls surgically removed at the 2nd Department of General and Oncological Surgery, Wroclaw Medical University between January 2008 and April 2014. The subsequent information, such as grade and histological type, location and size of tumor, lymph node status, hormonal (ER, PR) and menopausal status and patient’s age was gained from medical records.
Immunohistochemical analysis
All women had Patey’s conservative radical mastectomy. In regard to the recommendations of the Polish Union of Oncology, postoperative adjuvant therapy was performed. Immunohistochemical analysis was conducted according to the methods of Agrawal et al. 2016 (12). First, FFPE samples were sliced at 4 mm sections. Afterward, specimens underwent deparaffinization in two fresh xylene soaks, rehydration by using alcohol washes of decreasing concentrations (96%, 80%, 70%), and staining with hematoxylin-eosin. Then, slides were washed in distilled water, dehydrated through alcohols (70%, 80%, 96%), cleared in xylene and mounted in mounting medium. The H&E stained sections were prepared for the histopathologic examination. Following the histopathological analysis, further immunohistochemical staining of ARID1A antibody expression was evaluated. Prior to antibody staining, the epitope retrieval step was performed by boiling slides in a citric buffer [0.1M citric acid, 0.05% Tween 20 (pH 6.0); Sigma-Aldrich] for 8 minutes. To remove non-specific bindings and the staining from the background, specimens were incubated for 10 minutes with a peroxidase blocking reagent and, after washing, with protein block reagent for another 10 minutes. The tissues were incubated with primary antibodies and stored overnight at 4 °C [ARID1A Antibody (PSG3); sc-32761 dilution 1/100, Santa Cruz]. The next day, after PBS washing, slides were incubated for 15 minutes with secondary biotin and streptavidin-HRP-conjugated antibody. The immunoreactivity was visualized using 3,3-diaminobenzidine (DAB) method. Finally, the samples were counterstained with hematoxylin, dehydrated and cleared in xylene.
The BAF250a protein expression was presented as a percentage of tumor cells exhibiting a positive reaction. Two independent pathologists evaluated immunohistochemical reaction using light microscopes (Olympus BX51) and the results were agreed thereafter.
Statistics
The statistical analysis was performed by the use of Statistica v. 12 software (StatSoft, Inc., Tulsa, OK, USA). Expression values between normal (control) and cancer specimens were compared by the non-parametric Mann-Whitney U test and for the comparison between three or more groups, the Kruskal-Wallis test followed by Dunn’s multiple comparison test was conducted. P values lower than 0.05 (P<0.05) were considered as statistically significant.
Results
The clinicopathological features of a total of 119 female patients with primary invasive breast carcinoma are listed in Table 1. Patients’ age ranged between 29 and 84 years (mean: 59.0 years; SD: 12.9). The majority of the group consisted of women at postmenopausal age (89/119; 73.1%). The most common histological type of tumor was ductal carcinoma (99/119; 83.2%). The sizes of the tumors varied from 2 to 45 mm with an average of 18.5 mm (SD: 8.3). More than often (63/119; 52.9%) the right breast was affected by the tumor and almost half of the tumors (54/119; 45.4%) were localized in the upper outer quadrant of the breast. Among all of the 119 patients’ tumors were classified as pT1: 81 (68.1%), pT2: 37 (31.1%), and pT3: 1 (0.8%). Over half of the experimental group (59.7%) had no lymph node positivity at the time of surgery. None of the patients showed distant metastases.
Table 1
| Variable | Statistics |
|---|---|
| Age (at the time of diagnosis) | |
| M ± SD | 59.0±12.9 |
| Me (Q1; Q3) | 59 (48; 67) |
| Min to max | 29–84 |
| Age group | |
| Between 31 and 40 years old | 8 (6.7%) |
| Between 41 and 50 years old | 27 (22.7%) |
| Over 50 years old | 84 (70.6%) |
| Menopausal status | |
| Premenopause | 32 (26.9%) |
| Postmenopause | 87 (73.1%) |
| Diagnosis | |
| Ductal carcinoma | 99 (83.2%) |
| Lobular carcinoma in situ | 8 (6.7%) |
| Intraductal carcinoma | 4 (3.4%) |
| Other carcinoma | 8 (6.7%) |
| Tumor size (mm) | |
| M ± SD | 18.5 ± 8.3 |
| Me (Q1; Q3) | 18 (13; 23) |
| Min to max | 2–45 |
| Number of patients with primary tumor size: | |
| pT1 | 81 (68.1%) |
| pT2 | 37 (31.1%) |
| pT4 | 1 (0.8%) |
| Side | |
| Right breast | 63 (52.9%) |
| Left Breast | 56 (47.1%) |
| Primary tumor location | |
| Upper outer quadrant | 54 (45.4%) |
| Upper inner quadrant | 28 (23.5%) |
| Lower outer quadrant | 16 (13.4%) |
| Lower inner quadrant | 3 (2.5%) |
| Superior breast | 5 (4.2%) |
| Inferior breast | 2 (1.7%) |
| Outer breast | 1 (0.8%) |
| Inner breast | 1 (0.8%) |
| Central breast | 9 (7.6%) |
| Number of patients with metastatic lymph nodes | |
| pN0 | 71 (59.7%) |
| pN1 | 38 (31.9%) |
| pN2 | 7 (5.9%) |
| pN3 | 3 (2.5%) |
| Metastatic lymph nodes (metastasis) | |
| M ± SD | 1.8±4.7 |
| Me (Q1; Q3) | 0 (0; 2) |
| Min to max | 0–38 |
| Number of lymph nodes dissected during operation | |
| M ± SD | 13.2±5.8 |
| Me (Q1; Q3) | 13 (10; 17) |
| Min to max | 1–38 |
| Number of patients with distant metastases: | |
| pM0 | 119 (100.0%) |
M, average; SD, standard deviation; Me, median; Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
The mean value of BAF250a expression in the experimental group was higher (79.4%; SD: 21.1) than in healthy control (67.6%; SD: 18.1) and the difference was statistically significant (P=0.001; Mann-Whitney test; Figure 1). The characteristics of the tested marker expression are shown in Table 2.
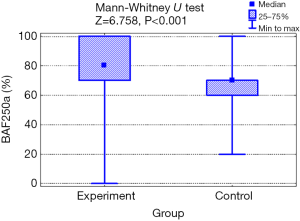
Table 2
| Expression BAF250a (%) | Test group (n=119) | Control group (n=92) | Test result |
|---|---|---|---|
| M ± SD | 79.4 ± 21.1 | 64.5 ± 14.9 | P=0.001 |
| Me (Q1; Q3) | 80 (70; 100) | 70 (60; 70) | |
| Min to max | 0–100 | 20–100 |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
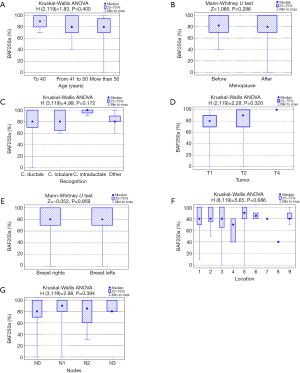
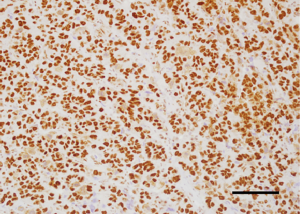
Analysis of the relationship between clinicopathological factors such as age, menopausal status, histological subtype of the tumor, size of the tumor, affected side of the body, location and lymph node metastases) and BAF250a expression shows no statistically significant correlation (P>0.05; Kruskal-Wallis/Mann-Whitney tests; Tables 3-9, Figure 2). Figure 3 presents the positive immunohistochemical staining for BAF250a in cells of invasive ductal breast carcinoma.
Table 3
| BAF250a (%) | Age group | Test result | ||
|---|---|---|---|---|
| Below 40 years old (n=8) | Between 41 and 50 years old (n=27) | Over 50 years old (n=84) | ||
| M ± SD | 88.8±12.5 | 80.6±19.3 | 78.1±22.2 | P=0.400 |
| Me (Q1; Q3) | 90 (80; 100) | 80 (70; 100) | 80 (70; 950) | |
| Min to max | 70–100 | 40–100 | 0–100 | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 4
| BAF250a (%) | Status | Test result | |
|---|---|---|---|
| Before menopause (n=32) | After menopause (n=87) | ||
| M ± SD | 83.3±17.1 | 77.9±22.3 | P=0.286 |
| Me (Q1; Q3) | 82.5 (70; 100) | 80 (70; 100) | |
| Min to max | 40–100 | 0–100 | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 5
| BAF250a (%) | Diagnosis | Test result | |||
|---|---|---|---|---|---|
| Ductal carcinoma (n=98) | Lobular carcinoma in situ (n=8) | Intraductal carcinoma (n=4) | Other carcinoma (n=9) | ||
| M ± SD | 78.1±22.1 | 81.3±17.3 | 97.5±5.0 | 83.3±12.2 | P=0.172 |
| Me (Q1; Q3) | 80 (70; 100) | 80 (65; 100) | 100 (95; 100) | 80 (80; 90) | |
| Min to max | 0–100 | 60–100 | 90–100 | 60–100 | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 6
| BAF250a (%) | Tumor | Test result | ||
|---|---|---|---|---|
| T1 (n=81) | T2 (n=37) | T4 (n=1) | ||
| M ± SD | 79.3±18.6 | 78.9±26.0 | 100 | P=0.320 |
| Me (Q1; Q3) | 80 (70; 90) | 90 (70; 100) | – | |
| Min to max | 0–100 | 0–100 | – | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 7
| BAF250a (%) | Side of body | Test result | |
|---|---|---|---|
| Right breast (n=63) | Left breast (n=56) | ||
| M ± SD | 79.0±22.0 | 79.9±20.2 | P=0.959 |
| Me (Q1; Q3) | 80 (70; 100) | 80 (70; 100) | |
| Min to max | 0–100 | 0–100 | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 8
| Tumor location | BAF250a (%) | |||
|---|---|---|---|---|
| N | M ± SD | Me (Q1; Q3) | Min to max | |
| Upper outer quadrant | 54 | 80.1±20.5 | 80 (70; 100) | 10–100 |
| Upper inner quadrant | 28 | 82.5±16.0 | 80 (75; 100) | 50–100 |
| Lower outer quadrant | 16 | 75.6±27.6 | 80 (65; 100) | 0–100 |
| Lower inner quadrant | 3 | 63.3±20.8 | 70 (40; 80) | 40–80 |
| Superior breast | 5 | 74.0±42.2 | 90 (80; 100) | 0–100 |
| Inferior breast | 2 | 85.0±7.1 | 85 (80; 90) | 80–90 |
| Outer breast | 1 | 80 | 80 | 80–80 |
| Inner breast | 1 | 40 | 40 | 40–40 |
| Central breast | 9 | 83.3±10.0 | 80 (80; 90) | 70–100 |
| Kruskal-Wallis test | P=0.686 | |||
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Table 9
| BAF250a (%) | Lymph node metastases | Test result | |||
|---|---|---|---|---|---|
| N0 (n=71) | N1 (n=38) | N2 (n=7) | N3 (n=1) | ||
| M ± SD | 77.2±21.8 | 82.9±19.4 | 79.3±26.2 | 86.7±11.5 | P=0.394 |
| Me (Q1; Q3) | 80 (70; 100) | 90 (80; 100) | 85 (60; 100) | 80 (80; 100) | |
| Min to max | 0–100 | 0–100 | 30–100 | 80–100 | |
M, average; SD, standard deviation; Me, median, Q1, lower quartile (25%); Q3, upper quartile (75%); Min, minimum value; Max, maximum value.
Discussion
Mammalian SWI/SNF is highly conservative and consists of 12–28 subunits (1). Within this class of complexes, two categories can be distinguished: (I) BAF (Brg/Brahma-associated factors) complexes consisting of ARID1A/B (AT-rich interactive domain containing protein 1A/B) subunits and (II) PBAF (polybromo-associated BAF) complexes consisting of ARID2 (AT-rich interactive domain containing protein 2) and PBMR1 (Polybromo 1) subunits (1,8). Chromatin regulatory machinery modifies nucleosome structure and modulates DNA accessibility to cellular mechanisms involving transcription, proliferation and DNA damage response (9).
BAF250a/ARID1A belongs to the family of DNA-binding proteins and regulates SWI/SNF chromatin remodeling complex activity in cell nucleus (13). According to Li and co-workers BAF250a affects in direct manner histone H2B using ubiquitination (14). Its expression is correlated with cell cycle and shows the highest level in G0/G1 phase and lowest during S, G2/M phases (15). Promoter-BAF250a cooperation is associated with cell cycle genes repression (16) and normal cell cycle arrest is compromised in cells with BAF250a depletion (17). As BAF250a/ARID1A plays a role in cell cycle repression, it is presumed to be a tumor suppressor based on loss-of-function mutational profiles observed in a broad variety of cancers (18). Furthermore, BAF250a/ARID1A as a potential tumor suppressor gene may contribute to carcinogenesis and cancer progression (19).
ARID1A mutations are not frequently reported in breast cancers. Mutated ARID1A is detected in approximately 4% of BCs. Copy number loss is more frequent and involves 13–35% of BC cases (4,20,21).
According to Zhang et al. in a variety of primary invasive BCs limited BAF250a, expression results from hypermethylation of ARID1A gene promotor (22). BAF250a limited expression shows a significant correlation with ARID1A gene hypermethylation in 86.4% of invasive ductal breast cancers (19). Yamamoto and co-workers reported that BAF250a subunit dysregulation may alter PI3K/Akt, the signaling pathway and this correlation potentially links low BAF250a expression with ovarian clear cell carcinoma (23).
According to Mamo et al. decreased ARID1A expression is potentially associated with higher aggressiveness of BCs (21). However, prognostic values of ARID1A gene mutation remain unclear. Kandoth and co-workers found that ARID1A mutations are not significant in BC (24). Contrarily, Zhao and co-workers in their study indicated ARID1A deletion as an independent prognostic marker in BC (25). Moreover, Lin et al. reported that ARID1A down-regulation was related to a poorer response to paclitaxel-based chemotherapy (26).
As it has been reported the SWI/SNF subunits expression differs significantly in various human cancer cell lines (27). Data concerning aberrant ARID1A mRNA/protein expression in breast cancer tissue as well as breast cancer cell lines is highly diversified. According to Wang et al. BAF250a activity is deficient in 10% BCs (28) and in contrast to other studies shown, even up to 64% (21).
An inconsistent body of evidence encouraged our research to investigate BAF250a expression in BC patients diagnosed with different types of primary invasive carcinomas. In contrast to our initial hypothesis and previous papers on a limited number of patients, our data based on a cohort of female patients show that ARID1A is significantly overexpressed in primary invasive breast cancer comparing to healthy control samples. Moreover, the expression is unrelated to age, menopausal status, lymph node status, tumor size and location, grade and histologic type of tumor, and hormonal status (ER, PR). The results do not confirm the expression pattern described by Zhao et al. and Lin et al. (25,26). These contrasting results may be explained by the recent findings of Sun et al. that gain of ARID1A function promoted initiation via enhanced oxidative stress, while loss of ARID1A during the later phases of tumor growth decreased DNA accessibility and inhibited transcription of genes associated with migration, invasion, and metastasis (18). We presume that ARID1A may play context-dependent tumor-promoting and tumor-suppressive roles in cancer. Further studies are warranted to explain the observed differences in the expression of this biomarker.
Acknowledgments
Funding: This work was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2422). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics approval (number: KB 132/2018) was obtained by the Bioethics Committee of the Wroclaw Medical University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Biegel JA, Busse TM, Weissman BE. SWI/SNF chromatin remodeling complexes and cancer. Am J Med Genet C Semin Med Genet 2014;166C:350-66. [Crossref] [PubMed]
- Feinberg AP. Epigenetic stochasticity, nuclear structure and cancer. The implications for medicine. J Intern Med 2014;276:5-11. [Crossref] [PubMed]
- Choi JD, Lee JS. Interplay between epigenetics and genetics in cancer. Genomics Inform 2013;11:164-73. [Crossref] [PubMed]
- Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat 2012;33:100-3. [Crossref] [PubMed]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011;11:481-92. [Crossref] [PubMed]
- Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBMR1 in renal carcinoma. Nature 2011;469:539-42. [Crossref] [PubMed]
- Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer. Mechanistic insights gained from human genomics. Sci Adv 2015;1:e1500447. [Crossref] [PubMed]
- Luchini C, Veronese N, Solmi M, et al. Prognostic role and implications of mutation status of tumor suppressor gene ARID1A in cancer: a systematic review and meta-analysis. Oncotarget 2015;6:39088-97. [Crossref] [PubMed]
- Xu G, Ghhangawala S, Cocco E, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet 2020;52:198-207. [Crossref] [PubMed]
- Takao C, Morikawa A, Ohkubo H, et al. Downregulation of ARID1A, a component of the SWI/SNF chromatin remodeling complex, in breast cancer. J Cancer 2017;8:1-8. [Crossref] [PubMed]
- Agrawal AK, Pielka E, Lipinski A, et al. Clinical validation of nuclear factor kappa B expression in invasive breast cancer. Tumour Biol 2018;40:1010428317750929. [Crossref] [PubMed]
- Agrawal A, Ziolkowski P, Grzebieniak Z, et al. Expression of Androgen Receptor in Estrogen Receptor–positive Breast Cancer. Appl Immunohistochem Mol Morphol 2016;24:550. [Crossref] [PubMed]
- Zhang X, Zhang Y, Yang Y, et al. Frequent low expression of chromatin remodeling gene ARID1A in breast cancer and its clinical significance. Cancer Epidemiol 2012;36:288-93. [Crossref] [PubMed]
- Li XS, Trojer P, Matsumura T, et al. Mammalian SWI/SNF – a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol 2010;30:1673-88. [Crossref] [PubMed]
- Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, et al. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res 2011;345:137-48. [Crossref] [PubMed]
- Nagl NG Jr, Wang X, Patsialou A, et al. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J 2007;26:752-63. [Crossref] [PubMed]
- Nagl NG Jr, Patsialou A, Haines DS, et al. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res 2005;65:9236-44. [Crossref] [PubMed]
- Sun X, Wang SC, Wei Y, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer Cell 2017;32:574-89.e6. [Crossref] [PubMed]
- Cho HD, Lee JE, Jung HY, et al. Loss of Tumor Suppressor ARID1A Protein Expression Correlates with Poor Prognosis in Patients with Primary Breast Cancer. J Breast Cancer 2015;18:339-46. [Crossref] [PubMed]
- Cornen S, Adelaide J, Bertucci F, et al. Mutations and deletions of ARID1A in breast tumors. Oncogene 2012;31:4255-6. [Crossref] [PubMed]
- Mamo A, Cavallone L, Tuzmen S, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene 2012;31:2090-100. [Crossref] [PubMed]
- Zhang X, Sun Q, Shan M, et al. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PLoS One 2013;8:e53931. [Crossref] [PubMed]
- Yamamoto S, Tsuda H, Takano M, et al. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol 2012;25:615-24. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Zhao J, Liu C, Zhao Z. ARID1A: a potential prognostic factor for breast cancer. Tumour Biol 2014;35:4813-9. [Crossref] [PubMed]
- Lin YF, Tseng IJ, Kuo CJ, et al. High‐level expression of ARID1A predicts a favourable outcome in triple‐negative breast cancer patients receiving paclitaxel‐based chemotherapy. J Cell Mol Med 2018;22:2458-68. [Crossref] [PubMed]
- Hughes AL, Owen-Hughes T. Deciphering Subunit-Specific Functions within SWI/SNF Complexes. Cell Rep 2017;18:2075-6. [Crossref] [PubMed]
- Wang X, Nagl NG, Wilsker D, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J 2004;383:319-25. [Crossref] [PubMed]


